Benoît Kornmann
@BenoitKornmann
Followers
1K
Following
460
Media
169
Statuses
952
@KornmannLab at @UniofOxford and @StHughsCollege
Oxford, England
Joined February 2016
Note to self: don't forget to pin a thoughtful tweet in my profile. .
0
0
6
(just quickly back on Twitter because I promised myself to also tweet about the not-so-rosy things in the job).
0
0
2
Terrible case of "esprit de l'escalier" (this is a thing, Google it). In the coach, back from defending my proposal in front of the panel,. and thinking of aaaaall the perfectly good answers to their questions. Missed them all, pretty much. 😣.
4
0
12
Nucleotide influences client binding. What's the mechanism?. Voilà. thanks for reading!.
0
0
1
This will spark more research!. The hydrophobic pocket is part of a Ca++ binding domain. What's the crosstalk?. We now can surgically disrupt client-Miro interaction without affecting other functions of client or Miro. What's the significance?.
1
0
0
In conclusion.-AlphaFold is a fantastic tool to revisit old knowledge.-Miro proteins are general purpose adaptors that recruit a host of client on mitochondria.-They all bind the same pocket (thus competitively).
1
0
0
Here we use AlphaFold to predict Miro interaction with 10 different clients. We then validate the predictions. -all clients use the same hydrophobic pocket in Miro to insert a L or F residue. -all have a similar enough motif that it allows us to identify a new family of clients.
1
0
0
This led to some confusion; how can such proteins bind so many different partners? Is it one large supacomplex or many different smaller ones?.What does Ca++ or nucleotide binding do to Miro activity, etc.?.
1
0
0
Instead, we found yeast Miro as part of an ER-mito tethering complex, the ERMES. Over the years, studies have additionally implicated Miro in mito attachment to actin, interorganelle lipid transport, mitophagy etc. In each study, (a) new binding partner(s) have been found.
1
0
0
Miro have originally been found in mammals and drosophila as important adaptors for microtubule motors. Yet, Miro are found in organisms (like yeast) that do not transport mitocohndria on microtubules. Therefore cytoskeletal adaptor cannot be their ancestral function.
1
0
0
Miro is a fascinating family of proteins.-They're ultraconserved in eukaryotes.-They harbour two GTPases domains and two Ca++-binding domains.-They're on mitochondrial surface. What else do we know about these?.
1
0
0
🚨New preprint Alert🚨. Shared Structural Features of Miro Binding Control Mitochondrial Homeostasis . If you want to get the gist follow the 🧵.
biorxiv.org
Miro proteins are universally conserved mitochondrial calcium-binding GTPases that regulate a multitude of mitochondrial processes, including transport, clearance and lipid trafficking. Miro binds a...
1
14
30
just stumbled upon a @eLife paper. Title seemed awesome but out of my field. couldn't know if it was a paper I needed to read ASAP or if it could wait. I just read the eLife assessment and.got much more info than just knowing the journal name. Get into the habit of doing that!.
1
1
5
Great talk by Ben Barres Spotlight awardee @AJKowaltowski !. With seminars like this, science has no problem reaching all corners of the globe. Journals' purpose isn't dissemination anymore, but evaluation. Time to endorse this and change the way we publish. @eLife.
Thank you to @AJKowaltowski for the great lecture, to @BenoitKornmann for the discussion on the new @eLife model, and to the 250+ participants!. See you again next month, for another @MITOtalk!.
0
3
7
Undisclosed conflict of interest . (why am I even trying to imitate the inimitable @OdedRechavi ?)
0
1
25
Not very active on twitter anymore but THIS deserves a shoutout 🤯.
nature.com
Nature - Culture and analysis of ‘Candidatus Lokiarchaeum ossiferum’—a member of the Asgard phylum—reveals an elaborate cell architecture with extensive membranous protrusions.
0
0
3
RT @NatMetabolism: #NatMetabPicks | In @NatureCellBio, the team led by @BenoitKornmann (@UniofOxford, @StHughsCollege) develop METALIC, a t….
nature.com
Nature Cell Biology - John Peter et al. develop METALIC (Mass tagging-Enabled TrAcking of Lipids In Cells), an approach to track interorganelle lipid flux in live cells using organelle-targeted...
0
9
0
RT @NatureCellBio: NEW ONLINE: @KornmannLab @ATJohnPeter and colleagues develop METALIC, an approach to track interorganelle lipid flux in….
nature.com
Nature Cell Biology - John Peter et al. develop METALIC (Mass tagging-Enabled TrAcking of Lipids In Cells), an approach to track interorganelle lipid flux in live cells using organelle-targeted...
0
4
0
Thanks James!.
This is so cool!! METALIC (Mass tagging-Enabled TrAcking of Lipids In Cells), a method to track interorganelle lipid flux inside cells, is used to study ERMES and Vps13 lipid transport function. Amazing @ATJohnPeter @BenoitKornmann! 🤩🔥👏
0
0
1
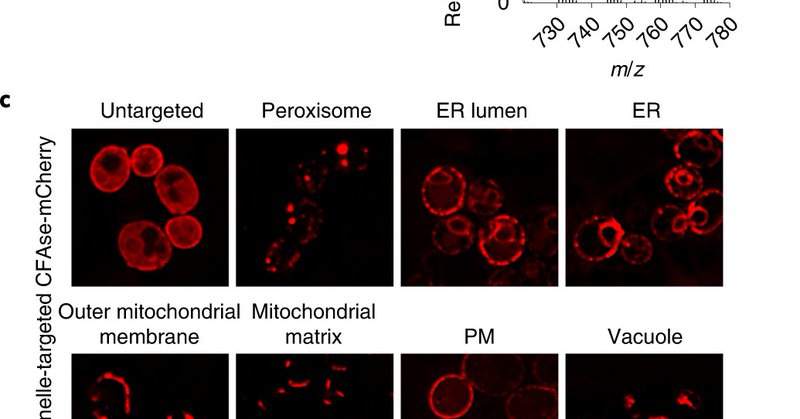
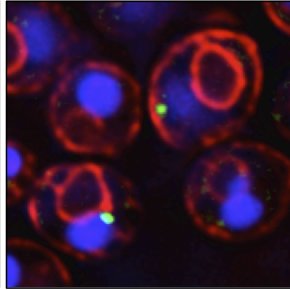
Our paper is out in @NatureCellBio .We present a new method to measure intracellular lipid trafficking in vivo and decipher the contributions of ERMES and Vps13-Mcp1 complexes in mitochondrial lipid supply.
nature.com
Nature Cell Biology - John Peter et al. develop METALIC (Mass tagging-Enabled TrAcking of Lipids In Cells), an approach to track interorganelle lipid flux in live cells using organelle-targeted...
8
83
328