Mujahid Ali, (Postdoctoral Researcher) PhD
@Ali_PhD87
Followers
62
Following
456
Media
1
Statuses
81
Study 3D chromatin organization and epigenetic mechanisms associated with learning and memory during critical periods# Institute of Neurosciences#Innsbruck
Innsbruck, Austria
Joined October 2018
preprint alert! Developmental regulation of Drosophila dosage compensation in a 3D genome context
biorxiv.org
Chromosomal dosage compensation (DC) restores the expression balance of X chromosome between sexes, and is realised in Drosophila by the coordination of enriched X-linked PionX, HAS sequence elements...
0
0
0
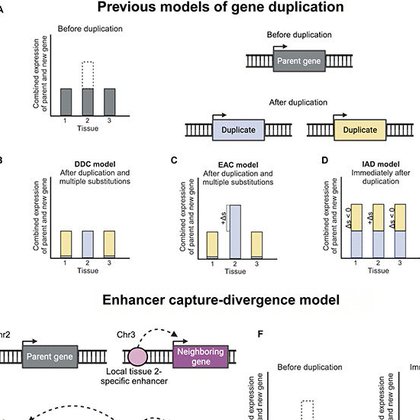
Happy to be part of this study that shows how proximity-based enhancer capture drives the evolution of distally duplicated genes, using Drosophila’s HP6/Umbrea as a model for this transformative genomic mechanism.
science.org
The 3D genome influences new gene evolution by altering gene expression via the general mechanism of enhancer capture-divergence.
0
0
0
RT @zhouqi1982: preatterned by active histone modifications, but zygotic genes by repressing modifications before zygotic genome activation….
0
1
0
RT @zhouqi1982: domesticated as enhancers. And specific but not housekeeping enhancers tend to coincide with 3D genome domain boundaries. W….
0
1
0
RT @zhouqi1982: We are excited to publish an modENCODE level epigenome data analyses for Dobzhansky's favourite Drosophila species D. pseud….
0
12
0
We are excited to share our latest work in @NatureComms! Our study maps the chromatin state transitions and highlights dynamic shift of chromatin states and regulatory regions(TABs,TEs and enhancers)during the development.#TEs,#enhancers,#sexchr,#3DGenome
0
0
11
RT @NatureRevGenet: Here, the authors synthesize recent insights into sex chromosomes from a variety of species to review their evolutionar….
0
7
0
RT @NatureRevGenet: Moreover, continued studies in model organisms with recently developed technologies have characterized the dynamics of….
0
1
0
RT @NatureRevGenet: Evolution and regulation of animal sex chromosomes #Review by Zexian Zhu, Lubna Younas (@Lubna9….
0
8
0
RT @zhouqi1982: My student Zexian, Lubna @Lubna99584738 and I @ZJU_China together wrote a review on @NatureRevGenet on evolution and regul….
0
17
0
RT @zhouqi1982: #NCOMTop25 Our paper on #sexchr #evolution of #worms has made it to top 25 most read articles of @NatureComms ! https://t.c….
0
3
0
RT @zhouqi1982: Our paper of studying the #sexchr evolution of the amazing #seahorse species is just out on @MolBi….
0
10
0
RT @UlrichTechnau: When teaching is fun: great seminar with motivated and interested students in our Master program Evolutionary Systems Bi….
0
5
0
RT @biorxiv_evobio: The Role of the 3-Dimensional Genome in New Gene Evolution #biorxiv_evobio.
0
4
0
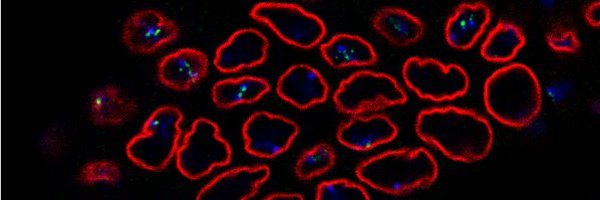
Excited to share our latest work on #Drosophila pseudoobscura #chromatin landscape.
0
1
3
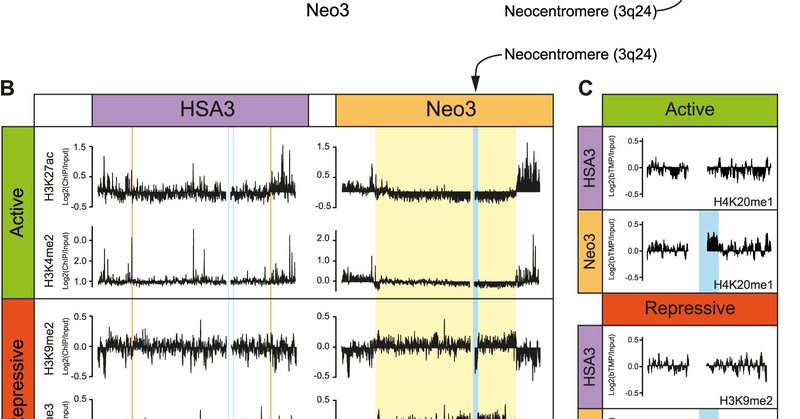
RT @chromatinlab: Check out our latest study on centromere chromatin structure out now in @NatureComms
nature.com
Nature Communications - In this study, using a human neocentromere as a model, the authors show that centromeres have a special chromatin structure. Centromere repositioning triggers...
0
17
0
RT @VDSEE_univie: Today @JingLiu74952792 PhD did an amazing job defending his thesis in front of @huguesrc, @ProfJennyGraves and Oleg Simak….
0
3
0
RT @boris_ctcf: 3D genome, on repeat: Higher-order folding principles of the heterochromatinized repetitive genome .
0
17
0
RT @KarstenRippe: A lot has been published on the big chunks of H3K9me2/3 heterochromatin. But how do the thousands of neglected small doma….
0
14
0