Karsten Rippe
@KarstenRippe
Followers
1K
Following
105
Media
56
Statuses
164
Joined November 2014
9/ 🤘 It's a long way to the top if you wanna rock 'n' roll. but @IsabelleSeufert rocked it with help of great PhD/Master's students @DKFZ & BioQuant, and valuable contributions from @AkisPapantonis, @jpmallm1, @s_anders_m & Petros Kolovos labs & funding by @spp2202_dfg. #ACDC.
2
0
11
8/ 🔥 We reconcile findings from previous sequencing and microscopy studies into the genome-wide AC/DC model of #transcription regulation, revealing two co-existing chromatin compartment architectures. For the full story, check out our bioRxiv preprint at
1
0
9
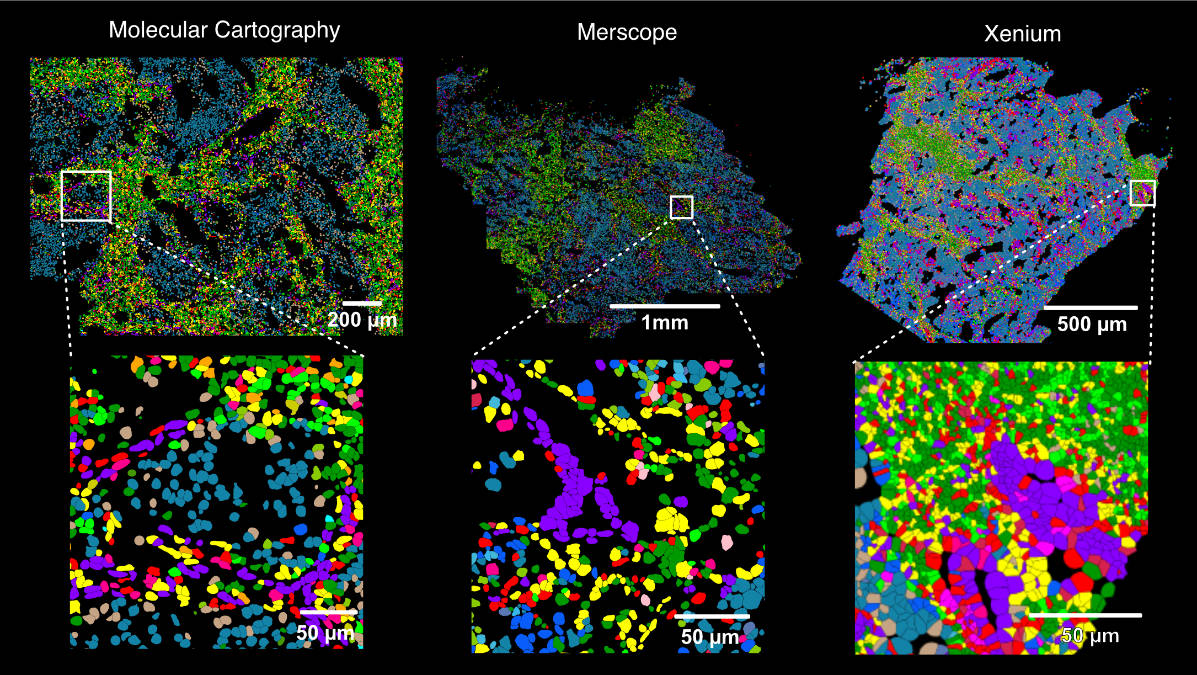
7/ 🌟 Our approach is applicable across other cellular systems and perturbations like interferon signaling of TF knock-out. Results improve with deeper #scATAC-seq coverage. We pushed that to 150k unique fragments/cell, but RWireX works well with 20-30k fragments/cell.
1
0
8
1/ 🤔 Do #chromatinlooping-mediated promoter-#enhancer interactions drive gene expression? Or is the local enrichment of transcription factors into hubs/#condensates the key factor? Our AC/DC model of gene regulation at bioRxiv provides a new view on this.
4
73
271
Great joint effort with @jpmallm1 from the Single Cell Open Lab @DKFZ and driven by Anne Rademacher, @alik_huseyn0v and Michele Bortolomeazzi with valuable contributions from our collaborators @KiTZ_HD and crucial funding from the MULTI-SPACE program of @ResearchLifesci.
0
1
5
RT @crc_1064: Registration is open! Join us in October for our quadrennial international #chromatin symposium in Munich. Hear and meet amaz….
0
14
0
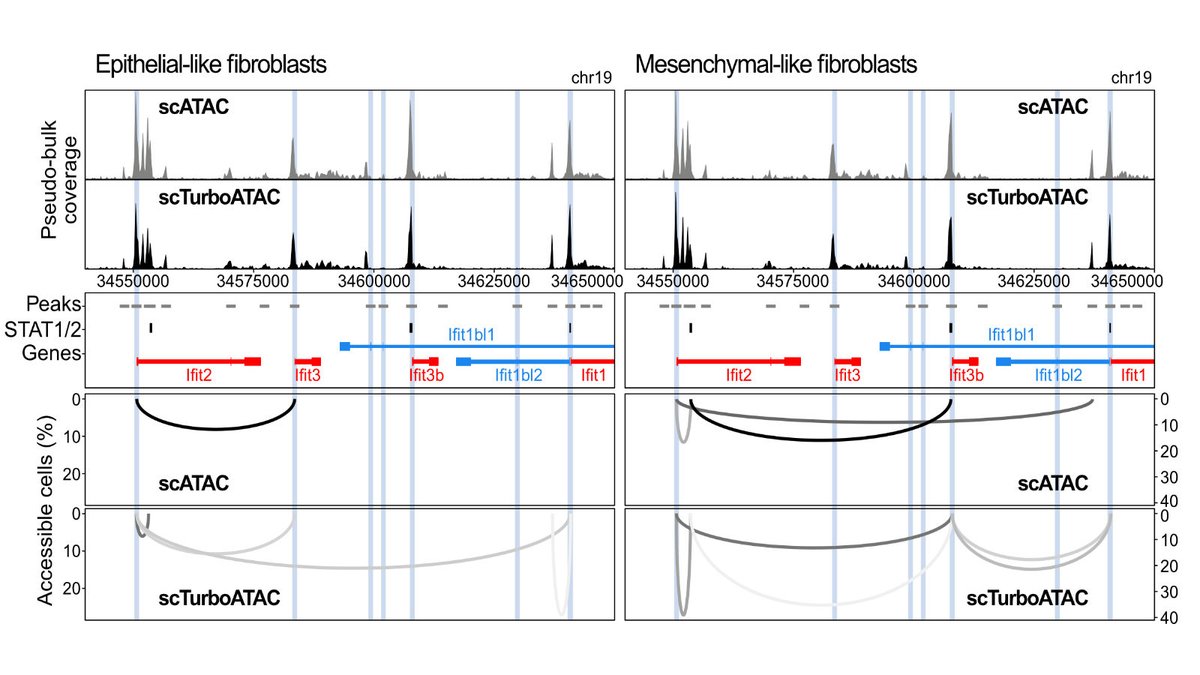
Enjoying the work with @IsabelleSeufert and @jpmallm1 to increase the coverage of scATAC-seq for co-accessibility analysis and other applications Our latest data are at 150k unique fragments/cell. Stay tuned.
1
6
24
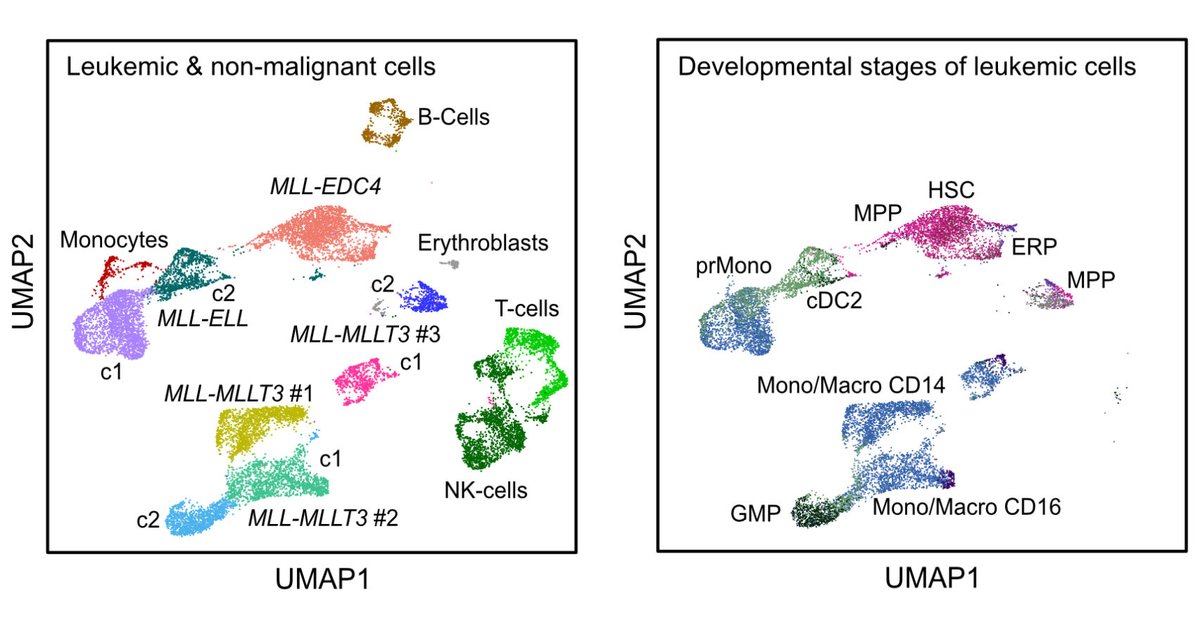
Our single cell analysis of MLL/KMT2A fusions in AML with Michael Lübbert's group @Uniklinik_Fr is now out in @BloodAdvances. It shows a progenitor-like phenotype of the novel MLL-EDC4 fusion. Free download link: Thanks to DFG FOR2674 for support.
0
8
37
RT @TeifLab: Our paper on nucleosome repositioning in CLL is now published in @genomeresearch! Led by @krissyvalp in our lab, collab with @….
0
40
0