Andreas-David Brunner
@ADBxMPIB
Followers
225
Following
444
Media
2
Statuses
130
Joined June 2019
RT @IdoAmitLab: Congratulations Ali @erturklab, Matthias @labs_mann, Fabian @fabian_theis and teams for this tour de force study, brilliant….
cell.com
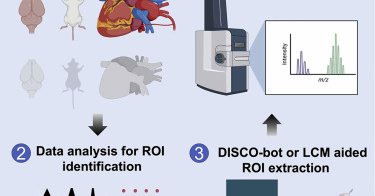
DISCO-MS and DISCO-bot allows for unbiased spatial proteome analysis of small tissue regions identified by panoptic imaging of large cleared samples, in silico reconstructions, and automated minima...
0
26
0
RT @Farida80168644: 📢@erturklab Spatial proteomics goes 3D🤯. Merging DISCO-clearing and proteomics to sample diseased tissues in their mil….
0
7
0
RT @erturklab: Spatial-omics goes 3D🤗! Out at Cell @CellCellPress 👉🏼 We developed DISCO-MS with @labs_mann, a spatial proteomics technology….
0
296
0
RT @fabian_theis: 1/n Single-cell data analysis is complex with many tools to choose from - but which ones work best and should be used whe….
0
406
0
RT @fabian_theis: Excited that our work on learning cell-cell communication in spatial transcriptomics is finally out @NatureBiotech! Led b….
0
157
0
RT @davidsebfischer: 1/n Cells in tissues are not independent. Yet, a lot of mathematical models deployed to explain gene expression of sin….
0
64
0
RT @RalserLab: Feeling nostalgic! tomorrow is our last day @TheCrick- Was privileged to supervise a brilliant team since 2013, 7 postdocs a….
0
9
0
RT @fabian_theis: Spatial transcriptomics is becoming increasingly popular, with differential studies coming in a lot. How should we compar….
0
19
0
RT @labs_mann: Together with @MolecularCell, we are celebrating their 25th anniversary by contributing a technology review highlighting cut….
cell.com
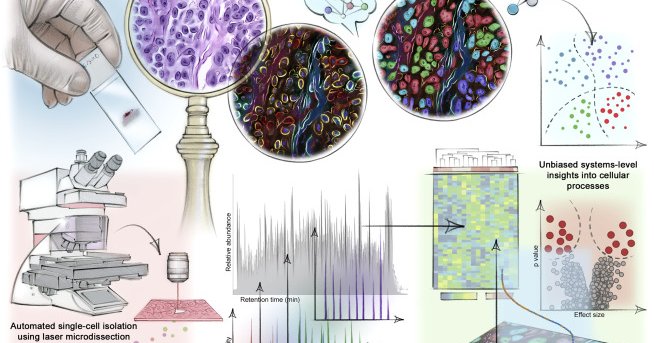
Mund et al. provide a perspective on single-cell proteomics and cell-type-resolved tissue proteomics and how they complement spatial transcriptomics. Together these technologies shed new light on...
0
37
0
RT @NanoPOTS_SCP: As single cell proteomics person, we always try to compare protein measurements to transcript, what if we can do both on….
biorxiv.org
Single-cell multiomics can provide comprehensive insights into gene regulatory networks, cellular diversity, and temporal dynamics. While tools for co-profiling the single-cell genome, transcriptome,...
0
55
0
Finally out in print - "Ultra-high sensitivity mass spectrometry quantifies single-cell proteome changes upon perturbation"! Thank you @MolSystBiol for selecting our article to cover the March issue and thank you to the whole team for this amazing project!
1
2
13
RT @MolSystBiol: 03/2022 Issue --> #SynBio, single-cell RNAseq of fly motoneurons, human kinase yeast array, #SynBi….
0
7
0
RT @LeonettiManuel: Our paper is out today in @ScienceMagazine, I’m stoked @OpenCellCZB !. Check out CRISPR taggin….
0
299
0
RT @LeonettiManuel: A tribute to team science! @labs_mann @czbiohub @kchev @ADBxMPIB @preethiragh @loicaroyer @marcoyannic @mattersOfLight….
0
1
0
RT @Karl_Mechtler: If you are a Ukrainian citizen and would like to look for a position in the field of proteomics, mass spectrometry or (b….
0
99
0
RT @dawafutisherpa: Our review on "How ends signal the end" is out in @MolecularCell. Here we discuss various aspects of numerous terminal….
0
18
0
RT @labs_mann: Our scalable, quantitative and ultrasensitive LC-MS based workflow for single cell proteome analysis 'one cell at a time' is….
0
74
0
RT @PastelBio: Ultra‐high sensitivity mass spectrometry quantifies single‐cell proteome changes upon perturbation | .
0
7
0