Yonghao Yu Lab - Hiring now!
@yu_yonghao
Followers
965
Following
1K
Media
18
Statuses
418
Professor at Columbia University. Passionate about (1) novel covalent protein modifications (eg., ADP-ribosylation) and (2) novel covalent inhibitors. views=own
Joined May 2019
Very excited to share our most research on novel PARP inhibitors. We identified a PARP1 trapping mechanism and the strategy to therapeutically target this pathway for BRCA mut cancers (to overcome PARPi resistance). This is a long-term collaboration with Dr. Tian Qin @QinLab
3
1
30
Paper published during postdoc just reached 1000 citations.
2
0
15
Very excited to share our new paper out in @Nature today, led by my amazing postdoc, @QZ_ChemSynBiol (now at @LifeAtPurdue) in collab w/ @themazelab (with Li & @MuirLab) showing a beautiful mechanism of monoamine modification swapping in neurons 🤩 https://t.co/HM1KYOQdtd
25
20
137
Excited to share our preprint reporting a completely new approach to study kinase signaling (ProKAS): "Proteomic Sensors for Quantitative, Multiplexed and Spatial Monitoring of Kinase Signaling" We applied it to DDR kinases. Please share. https://t.co/QNIhKb71nL
2
10
46
Check out this thoughtful Preview from Billy on our recent manuscript in Molecular Cell. Thanks Billy!
Pioneer TFs have long been considered "undruggable" 💊 🧬 However, @scrippsresearch teams led by Ben Cravatt and @Michael_A_Erb have elegantly developed a first-in-class ligand for the pioneer TF FOXA1. In @MolecularCell, we commented on this groundbreaking work:
1
4
41
How does DNA damage signaling suppress chromosomal rearrangements? Check our recent paper just out in EMBO Journal. Multi-step control of homologous recombination via Mec1/ATR suppresses chromosomal rearrangements | https://t.co/6iNQe7T6bJ
5
26
78
(4) and finally, our recent work on the identification of lineage specific response as a novel synthetic lethality mechanism for PARP inhibitors. The talk has been uploaded to YouTube, and can be viewed from the this link.
0
0
0
In this seminar, I presented our work on (1) PARylation proteomics, with a focus on PARylation on Asp and Glu residues. (2) PARP PROTAC as a means to decouple PARP inhibition vs PARP trapping. (3) the novel RNF114/nimbolide pathway as a novel mechanism to regulate PARP trapping
1
0
1
Our seminar at the "SocialDNAing" seminar series describing our recent work on mechanism of action studies of PARP inhibitors. Thanks @zha_lab again for the invitation. https://t.co/oQX6X9MLXn
1
0
4
KUDOS to the first author, Dr. Chiho Kim, for leading this extraordinary study. I would also like to thank our wonderful collaborators, Drs. John Minna, Ben Drapkin, Han Liang, Ling Cai. PARP inhibitiors for SCLC!
0
0
0
The biochemical effects of PARPi (e.g., PARP trapping and DNA damage) are general, regardless of cellular contexts. However, cells respond to this signal differently. Expression of lineage specific oncoproteins could be a novel synthetic leathality mechanism for PARP inhibitiors.
1
0
0
In contrast, in SCLC cells that do not express Huwe1, ASCL1 is not degraded, and there is no cell death. As a result, Huwe1 expression could be a predictive biomarker for the therapeutic efficacy of PARPi in SCLC.
1
0
0
Interestingly, we found that even though PARPi treatment induces an universal DNA damage signal in all SCLC cell lines tested, these various SCLC cells respond to this DNA damage signal differently. In cells expressing Huwe1, ASCL1 is robustly degraded, leading to cell death.
1
0
0
How ASCL1 gets degraded in response to genotoxic stress? We found that a DNA damage-sensitive E3 ubiquitin ligase, Huwe1, is critically involved in this process. ASCL1 is a substrate of Huwe1, and Huwe1 targets ASCL1 for degradation under genotoxic conditions.
1
0
0
Importantly, we found that ASCL1 is robustly degraded upon the treatment of not only PARPi, but also clinically relevant chemotherapeutic agents. This is an important discovery because of the well established sensitivity of SCLC to chemo.
1
0
0
Among the down regulated proteins, we were particularly intrigued by ASCL1, which is a lineage specific oncoprotein for SCLC. ASCL1 plays an essential role in maintaining the neuroendocrine differentiation states of SCLC. Furthermore, ASCL1 is a key survival factor for SCLC.
1
0
0
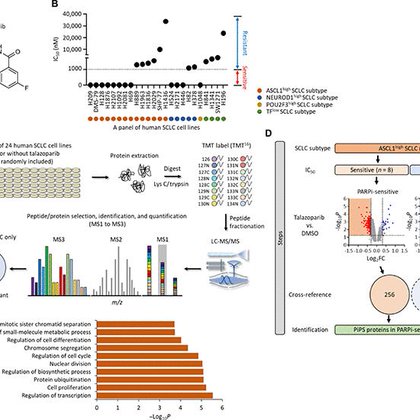
We performed large scale quantitative proteomics experiments to characterize how the proteomes of these SCLC cell lines respond to Talazoparib. We found that talazoparib-sensistive SCLC cells undergo a major remodeling of their proteomes in response to Talazoparib treatment.
1
0
0
Small cell lung cancer (SCLC) is a highly malignant and recalcitrant cancer. In this paper, we assembled a large panel of 24 SCLC cell lines that represent the major SCLC subtypes, I.e., ASCL1+, NeuroD1+, Pou2F3+, etc.
1
0
0
PARP1 as a therapeutic target for BRCAmut cancers has been well established. However, PARPi are increasingly being used for cancers without BRCA mutations. In this new publication, we sought to understand the mechanism of action of PARPi in SCLC.
science.org
Degradation of lineage-specific oncoproteins is a unique mechanism that drives PARPi sensitivity in small cell lung cancer.
1
2
10
SH2 domains thought of as "undruggable" - A News & Views by Oliver Hantschel highlights recent work from @alessiociulli lab @NatureComms on an inhibitor of the SH2 domain of E3 ligase SOCS2. Read it here https://t.co/7hnBD7AbR0 and the original article https://t.co/qdNdTo1XPx
0
9
75
See our most recent news&views article together with @QinLab in @NatureSynthesis on the discovery of the nimbolide-RNF114-PARP1 trapping pathway.
0
1
1