The Maze Lab
@themazelab
Followers
2K
Following
509
Media
19
Statuses
279
Understanding the complex interplay between chromatin regulatory mechanisms and neuronal plasticity. @icahnmountsinai @Sinaibrain @HHMINEWS
Mount Sinai-HHMI/Manhattan, NY
Joined January 2017
I am absolutely humbled and honored to have been selected as a new HHMI Investigator. A huge shout-out to all of my brilliant lab members (past and present), outstanding collaborators and incredible mentors along the way who made this a possibility!!
HHMI News: We are proud to announce our 33 new #HHMIInvestigators. These individuals have the potential to make transformative discoveries over time and we look forward to seeing where their ideas lead them.
22
8
228
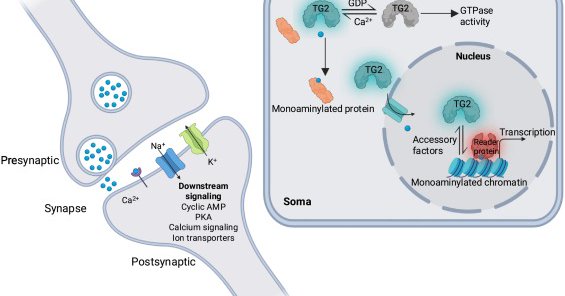
Reimagining biogenic amine signaling in the brain and beyond: Trends in Neurosciences
cell.com
Monoaminergic neurotransmission has long been recognized as essential for the development, maintenance, and plasticity of the nervous system, with classical models defining serotonin, dopamine, and...
0
4
16
🎉CONGRATULATIONS to the #MazeLab's @BenjaminWeekley who has received an @NIH F32 this year from @NIMHgov to study "Mechanisms of Histone Serotonylation and MLL5 in Coordinating the #Epigenome during #Neurodevelopment". Learn More about Dr. Weekley's work👉 https://t.co/i8mQIw4jqx
1
2
8
The One Mind Rising Star Awards were created to fuel bold ideas that improve lives, and 2017 Awardee Ian Maze, PhD, is a powerful example of that vision in action. Congratulations to Dr. @ian_maze2 on receiving the ACNP Daniel H. Efron Research Award for his groundbreaking work
0
2
6
So incredibly honored to be one of this year’s recipients of the @ACNPorg Daniel H. Efron Research Award, along with my friend and colleague @TheErinCalipari . Shout out to all my amazing current and former lab members (and our outstanding collaborators) who made this a reality!
Congratulations to Ian Maze, Ph.D. and Erin Calipari, Ph.D. for being awarded the Daniel H. Efron Research Award. 🎉#ACNP2026
0
1
10
Calling all rising juniors & seniors: Interested in biological or biomedical research? Apply now for our ’26 Summer Undergraduate Research Experience Program! Nine weeks, hands-on research, & mentorship from some of the nation’s top scientists — learn more:
1
104
173
New preprint alert!! In our last work we have found that Tissue stiffening primes migratory cell states by allowing histone serotonylation! #biomechanics #xenopus #chromatin #neuralcrest #devbio #cellmigration #frogpower
https://t.co/r5miQldHxh
biorxiv.org
Collective cell migration (CCM) is pivotal in several biological contexts, and posttranslational modifications of histones are essential to initiate this process[1][1]–[3][2]. Here, we show that a...
1
9
28
Thrilled to announce the upcoming Center for Epigenetic Research Symposium @MSKCancerCenter on Nov 17! 🎉 A full day of cutting-edge science featuring stellar external speakers & our outstanding trainees. Registration is now open! Join us in NYC or virtually from anywhere! link⬇️
1
5
27
So excited my doctorate work (and this excellent Preview by Yuhui Cui) is finally out @Neuron! We used human MDD profiling + mouse manipulation studies to identify an epigenetic regulator linking OFC astrocyte reactivity -> neuronal hyperexcitability -> stress susceptibility.
2
16
95
many amazing collaborators, without whom, none of this would have been possible. Hope you all enjoy!
0
0
5
But I couldn’t be more excited to finally have this work published. Also, a huge thanks to our incredibly helpful Editor, Dr. Ted Dobie, and our insightful Reviewers at Neuron, who truly demonstrated what scientific review should look like. Finally, a huge thanks to our
1
0
6
This project, which robustly identifies a novel astrocytic regulator of Major-depressive-disorder associated stress susceptibility represents a monumental effort by one of the most talented young scientists that I have ever known. Sasha will follow up with summaries of the work
1
0
4
So incredibly excited to be able to officially highlight the outstanding work of @SashaLFulton, a previous superstar PhD student in my lab and now a superstar postdoc in the @IshmailSaboor lab at Columbia (also an @HHMINEWS Hanna H. Gray Fellow), now out at @NeuroCellPress !!
Online now: Major-depressive-disorder-associated dysregulation of ZBTB7A in orbitofrontal cortex promotes astrocyte-mediated stress susceptibility https://t.co/WhWwIxQfyb
5
19
79
.@SinaiBrain & @IcahnMountSinai congratulate @EricJNestler for his election to @theNASciences of the USA. This great honor recognizes Dr Nestler’s cutting-edge work on the molecular basis of drug addiction & depression & on advancing the field of psychiatry into the molecular era
We are thrilled to announce the election of 120 members and 30 international members to the National Academy of Sciences in recognition of their distinguished and continued achievements in original research. Congratulations to our new #NASmembers and welcome to the Academy! 🎊
2
11
56
How do the brain & body communicate w/ each other? Our thoughts & emotions often feel in sync w/ physical sensations in our bodies. Why? Find out, CHECK OUT @RasikaIyer22's blog, dropped by MiNDS @SinaiNeuro this morning! #BrainAwarenessWeek #BrainWeek💓 https://t.co/uzkME0Ifwt
0
4
4
Today we're thrilled to announce our 2024 #HannaGrayFellows! Please join us in welcoming and celebrating these outstanding early career scientists! This cohort includes scientists working in research areas ranging from treatment-resistant cancers to sleep dysregulation to how
5
53
164
It was such a pleasure collaborating with you on this project, Qingfei! As they say, team work makes the dream work, and what an outstanding team this was 😎 here’s to many more!
So glad to see our paper out @Nature. I want to express my sincere thanks to my mentor @Yael_David__ , collaborators @themazelab and Haitao Li (who is also my undergraduate course teacher), and dear colleagues/friends. This could not happen without their guidance and support🩷.
1
1
11
And here is the beautifully written News & Views on our new paper from @debo_Astrocyte and Tatiana Kutateladze. Thank you both! https://t.co/dys66or82d
nature.com
Nature - Exchanger enzyme modifies histones in histamine-releasing neurons.
1
6
49
Massive thanks to all authors, including @QZ_ChemSynBiol @BenjaminWeekley @DavidVinson93 @RyanBastle Shuai Zhao and many many more on getting this across the finish line!
0
0
8
Using a viral approach to disrupt H3 monoaminylations in TMN, we significantly ablated circadian gene expression in this region. This matched behavioral phenotypes indicating significantly disrupted circadian entrainment in mice.
1
1
4
Through analysis, we show WDR5 binding is primarily at CLOCK/BMAL1 target genes, and binding was severely diminished at ZT20 when H3Q5his enrichment is high & H3Q5ser is low. This matches work showing MLL1 & CLOCK physically interact [Paulo Sassone-Corsi’s group (PMID 21113167)].
1
1
4