Vinoth
@vinoth46m
Followers
43
Following
100
Media
5
Statuses
18
PhD student @IITbombay
Mumbai, India
Joined October 2021
Our preprint, "Interplay between cohesin kinetics and polymer relaxation modulates chromatin-domain structure and dynamics," is now online. https://t.co/NqJQDJ6Dh7 We study how the interplay between extrusion and polymer relaxation affects chromatin structure & dynamics. (1/5)
5
9
43
1/n: There are some academic papers that are so brilliantly and so accessibly written and so universal in scope that they transcend disciplines and stand as timeless testaments to both great thinking and great writing. Here's a short personal selection:
98
1K
8K
Our latest paper to wrap up the year. We propose that intersegmental jumps that leverage looped conformations of chromatin can lead to efficient search. Read the preprint at: https://t.co/pqmNic2gl0 Led by @DuttaShuvadip & Adarshkrishnan. Fantastic collab. with @ranjithpa (1/4)
biorxiv.org
We investigate the role of compaction of chromatin domains in modulating search kinetics of proteins. Collapsed conformations of chromatin, characterised by long loops which bring distant regions of...
2
10
32
Chromatin compaction increases the length of spreading. Also, single-cell modification profiles need not be similar to the averaged ChIP-seq data. (5/5)
0
0
3
Writer enzymes, along with reader/writer enzymes can give rise to confined modification domains without any nucleation point. (4/5)
1
0
2
Local enzyme concentration controls the spreading of histone modifications (3/5)
1
0
2
The diffusion and the degradation of the enzymes set a length scale for the size of the modification domain. In a highly crowded nucleus where the diffusion constant is several orders of magnitude greater than water, this effect can be significant. (2/5)
1
0
2
What is the role of diffusion of enzymes/other constituents in the spreading of histone modifications? We simulated a model with diffusion and reaction of reader/writer enzymes to explain the spreading of histone modifications. https://t.co/ccPpior8zO (1/5)
1
10
41
@iitb4justice stands in solidarity with senior PhD scholars protesting against hostel shifting policy abruptly enforced on the batches severely affected by the pandemic.
2
60
57
Our work is out in @NatureComms. Read the paper if you want a quantitative description of chromatin polymer or plan to simulate chromatin. We answer some fundamental questions. Details below. @sangramjkadam @KiranKumari_ @vinoth46m @DuttaShuvadip @mithunmittir (1/10)
30
34
264
@Bhambid_Manasi 's work is out in The Protein Journal! She, @vishakhadey and @SujataWalunj show that T. gondii Importin alpha exhibits weak auto-inhibition. What are the implications of this for the biology of the parasite? Cool answers in the pipeline!
1
4
11
Our latest work in @PNASNews where we discuss how evolution selects sticky sequences in proteins to promote self-assembly & uses phosphorylation to safeguard against the dynamic arrest of these assemblies https://t.co/xpqx5mbijF
@ShakhnovichArmy @PouriaDasmeh Seth Furniss
pnas.org
Assemblies of multivalent RNA-binding protein fused in sarcoma (FUS) can exist in the functional liquid-like state as well as less dynamic and pote...
4
6
25
The #Mechanobiology workshop was organised by @BidishaSinha8, #PramodPullarkat, #VenkatesanIyer, #DeepakSinha & #SriRamaKotiAinavarapu & featured: - Techniques like Atomic Force Microscopy & optical/mechanical tweezers - As well as hands-on training sessions on optical trapping!
1
1
4
Chromatin is modelled as a bead-spring polymer. But how big is(physical size) a 10kb or 100kb chromatin bead? How flexible/bendable is chromatin? Starting nucleosome resolution, we coarse-grain chromatin & predict scale-dependent properties https://t.co/R6O3HExJZo
@sangramjkadam
biorxiv.org
Simulating chromatin is crucial for predicting genome organization and dynamics. Even though coarse-grained bead-spring polymer models are commonly used to describe chromatin, the relevant bead...
3
10
50
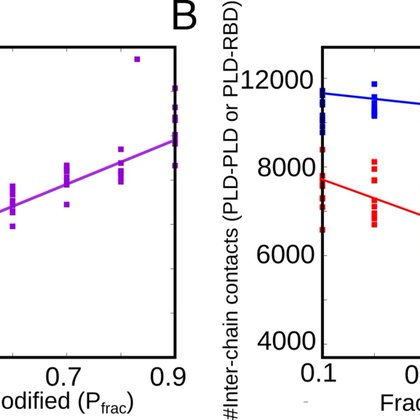
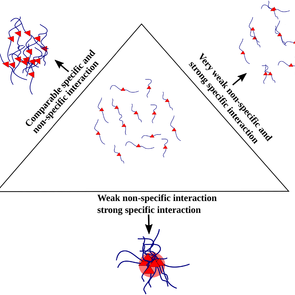
Our work in @PLOSCompBiol. Check here: https://t.co/FqBitPy15B. We were curious that, In the crowded cellular milieu, the process of biomolecules binding to specific interaction partners to carry out a function is non-trivial. (1/3)
journals.plos.org
Author summary Biological function relies on the ability of biomolecules to bind to specific interaction partners. In the crowded cellular milieu, the process of biomolecules binding to specific...
3
8
24
.@MargotRiggi and I are excited to release "Phase Separation 101," a visual resource for undergrad teaching. Check it out here: https://t.co/gTP4KZyqfi and many thanks to the researchers and educators who gave us their invaluable feedback! A short preview below:
29
362
1K
Every time we (TA's) receive a mail 3 to 4days prior that exams will be held and we have to proctor for the same. A Series of meetings and mock exams will be held in that window which totally messes up our work schedule (YES WE WORK!). The TA's hardly have a choice of their own.
1
1
9