tobiasgensch
@tobiasgensch
Followers
446
Following
256
Media
2
Statuses
77
Chemist in the Digital Age
Berlin
Joined April 2019
While I'm at it, my postdoc @Iris_JYG recently managed to capture an image of an elusive active catalytic species 😻
0
1
26
this morning one of my students casually dropped this in a presentation and I lost it 😂😂
16
79
920
Check out our first paper now published in @angew_chem, in which my group used machine learning tools to tailor our YPhos ligands for Hiyama couplings. Kudos to @JulianLoeff and the team as well as @tobiasgensch and the Gooßen group for a great collaboration.
Computer-Driven Development of Ylide Functionalized Phosphines for Palladium-Catalyzed Hiyama Couplings (Goossen) @SolvationSci @ViktoriaGessner @ruhrunibochum #OpenAccess thanks to #ProjektDEAL
https://t.co/a6VfEHYEkB
3
6
67
Very happy how our collaboration with @MilliporeSigma and @Sigman_Lab turned out! We designed a screening set for phosphine ligands using our kraken descriptor database with the idea to fully sample ligand chemical space and quickly obtain hits: https://t.co/xmY63zhJ3O
pubs.acs.org
In reaction discovery, the search space of discrete reaction parameters such as catalyst structure is often not explored systematically. We have developed a tool set to aid the search of optimal...
3
2
67
Ah wait...UniSysCat group leader @tobiasgensch et al. already got ahead of us! Check out the latest #research highlight at https://t.co/8w7zSTFFB0
#scicomm #wisskomm #catalysis #chemtwitter #sciencenews #MachineLearning
0
4
7
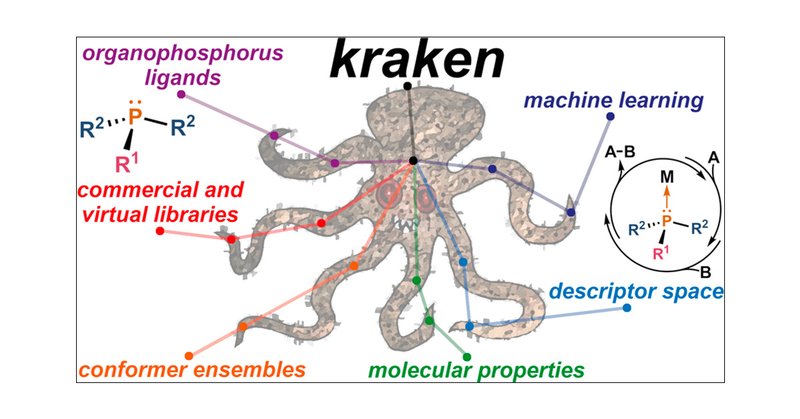
Kraken found a home in @J_A_C_S! It was an immense pleasure to work on this massive tour de force with @tobiasgensch @P_Friederich @robpollice @kjelljorner @akshat_ai @TheophileGaudin with guidance from @Sigman_Lab and the world-famous @A_Aspuru_Guzik!
Congrats to the team @tobiasgensch @gabepgomes @A_Aspuru_Guzik @robpollice and many others for building a unique, ML-enabled phosphine descriptor database!
7
8
105
Congrats to the team @tobiasgensch @gabepgomes @A_Aspuru_Guzik @robpollice and many others for building a unique, ML-enabled phosphine descriptor database!
pubs.acs.org
The design of molecular catalysts typically involves reconciling multiple conflicting property requirements, largely relying on human intuition and local structural searches. However, the vast number...
2
22
138
‘Accurately predicting the speciation of a catalyst would require precise knowledge of all the species that can be formed under given conditions and their relative energies’ – A new approach can identify novel ligands using only 5 experimental data points https://t.co/6RlXyUL9RU
chemistryworld.com
The approach was able to identify phosphine ligands that may form dinuclear palladium(I) complexes using only five experimental data points
0
9
10
HTE-data analysis enables the investigation of a new Pd-catalyzed electrophilic cyanation of aryl boronic acids @Sigman_Lab, @CoperetGroup, @tobiasgensch, @ETH_DCHAB, @likat_rostock @TUBerlin @UtahChemistry #openaccess
https://t.co/0ELkZ0dMID
0
6
19
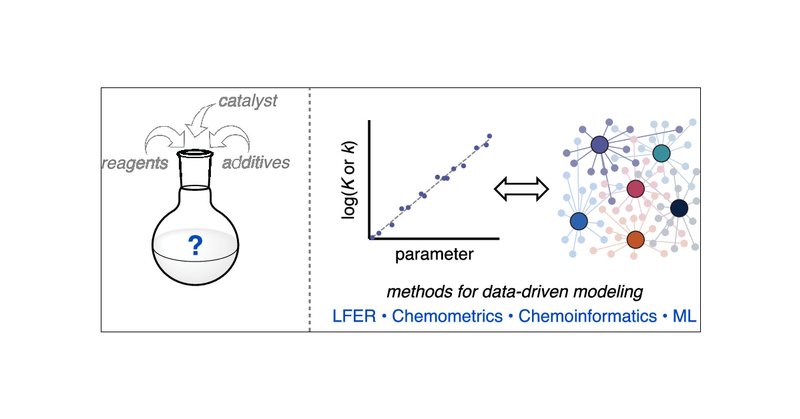
Really happy that our historical perspective on data-driven (physical) organic chemistry is now online! I had a lot of fun working with all these awesome people🙃
pubs.acs.org
Organic chemistry is replete with complex relationships: for example, how a reactant’s structure relates to the resulting product formed; how reaction conditions relate to yield; how a catalyst’s...
0
8
51
In @ScienceMagazine this week, @Sigman_Lab joins forces with Abby Doyle's group to predict when 2 of a given phosphine ligand bind to Ni or Pd
science.org
A descriptor predicts whether nickel and palladium are likely to coordinate to one or two of a given phosphine in catalysis.
2
26
138
Attending the #ACSFall2021? Please join us to discuss recent advances in Process Research and Development in Crop Protection with @corteva @Syngenta @Bayer4Crops @CornellaLab @AdrGomezSuarez @tobiasgensch @megnsium
0
5
13
In addition to the case studies shown in the manuscript above, @chemchristensen and @KingstonCian have already used this principle of designed phosphine screening sets in "real" projects: https://t.co/MQwFp4Wn77
https://t.co/hYLpcHrVfi
nature.com
Communications Chemistry - An automated closed-loop system optimizes a stereoselective Suzuki-Miyaura reaction using a machine learning algorithm that incorporates unbiased and categorical process...
0
2
9
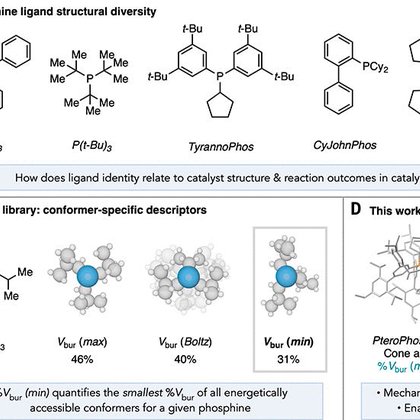
PHOSS is a 32-ligand-set that efficiently and completely probes the phosphine chemical space as represented by our kraken library. Sigma sells the ligands and access to a suggestions for (hopefully better) ligands based on the results obtained with PHOSS
1
4
11
As my part in the phosphine saga with @Sigman_Lab draws to an end, I am very proud to present our work on the design of ligand screening sets in collaboration with @MilliporeSigma @SigmaAldrich:
1
6
32
Heavy Metal(s)!🎸🤟 Today we are celebrating the birthday of our groups own Rockstar Tristan by showing some support for the new single of his band! If you like some good old melodic death metal while finishing the last columns of the day, check it out: https://t.co/9hJ210W1Uk
1
6
61
I absolutely love this work by Abby and her team! This addresses issues with the design of substrate scope tables that I have been thinking about myself for years, hopefully leading to more meaningful scope tables and less hiding of uncomfortable results(?)
Congrats to the Doyle lab - using #datascience to guide Ni/Photoredox-Catalyzed Cross-Couplings - now online @ChemRxiv
https://t.co/09HtvfLOPz
0
1
10
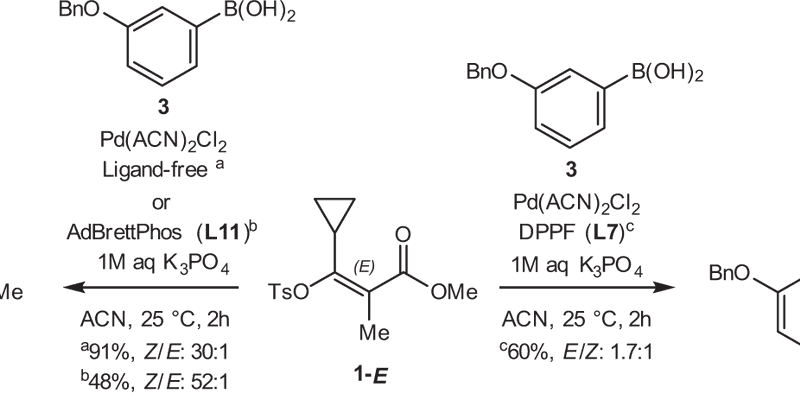
Here comes the next chapter in the phosphine saga and a fun little return to electrophilic cyanation reagents for me https://t.co/R8LdspfjaB I enjoyed working with Jordan @CoperetGroup and all the others, trying to make sense of this really nice ligand screening 🤖
0
1
16
A proud alumnus of @Sigman_Lab, @GloriusGroup, & @NSF_CCAS, Tobias Gensch (@tobiasgensch) is all set to take the @VISTA_chem virtual stage on Monday, July 19th, 4.40 PM CEST to tell his story on phosphine ligands! Join us to catch Tobias live🔥 @thiemechemistry @KathrinUlbrich
0
14
37