Wei Li
@superweili
Followers
809
Following
18
Media
1
Statuses
27
Professor of Bioinformatics at University of California, Irvine
Irvine, CA
Joined July 2010
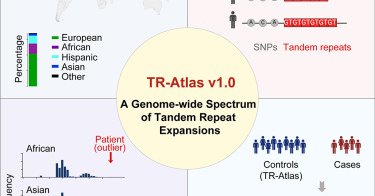
RT @CellCellPress: Now online! A genome-wide spectrum of tandem repeat expansions in 338,963 humans
cell.com
The Tandem Repeat Aggregation Atlas (TR-Atlas) is a biobank-scale reference of 0.86 million TRs derived from 338,963 whole-genome sequencing (WGS) samples of diverse ancestries.
0
16
0
Our new paper @NatureComms performed the first Alternative polyadenylation transcriptome-wide association study for 11 brain disorders with 17,300 RNA-seq samples. We identified 354 novel APA-linked risk genes including ATXN3 for ALS.
8
32
153
RT @jsb_ucla: Ultimately, adjusting hidden covariates or not examines different DE effects (conditional vs marginal). But regardless of thi….
0
4
0
RT @jsb_ucla: @borishej @mikelove @KasperDHansen @timtriche @andrewejaffe @lpachter @RodThiebaut @denis_agniel We thank @borishej et al for….
0
3
0
RT @GenomeBiology: Study of false positives when calling differentially expressed genes, from @YM123411, @ge_xinzhou, @FanglueP, @superweil….
0
68
0
RT @LeiLi_bioinfo: Population-scale genetic control of alternative polyadenylation and its association with human diseases..
0
2
0
Our new scDaPars tool for single-cell alternative polyadenylation is published .@genomeresearch.
1
7
39
RT @NatureGenet: 💥An atlas of alternative polyadenylation quantitative trait loci contributing to complex trait and disease heritability |….
nature.com
Nature Genetics - A multi-tissue atlas of alternative polyadenylation (APA) quantitative trait loci (3′aQTLs) identifies approximately 0.4 million common genetic variants associated with the...
0
8
0
RT @XShirleyLiu: Congratulations to @mourisl on the great improvement on the TRUST algorithm!
github.com
TCR and BCR assembly from RNA-seq data. Contribute to liulab-dfci/TRUST4 development by creating an account on GitHub.
0
1
0
Super excited to share our recent work on 3′UTR alternative polyadenylation (APA) quantitative trait loci (3′aQTLs), which can explain ~16.1% GWAS SNPs and are largely distinct from eQTLs. Wonderful collaborations with @Wagnerlab_RNA @LeiLi_bioinfo .
7
46
195
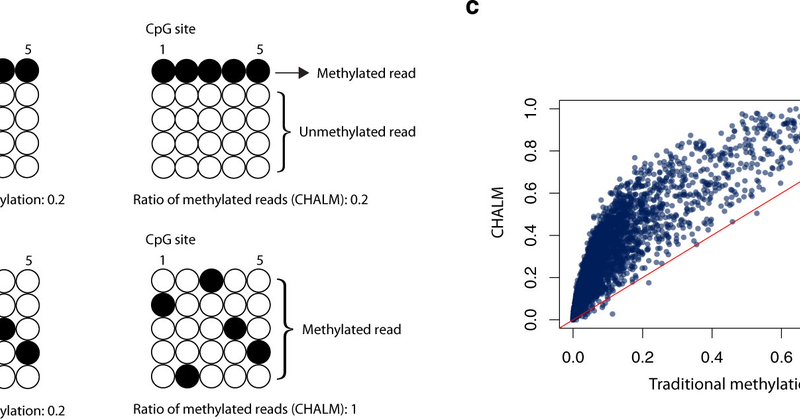
Excited to share our new paper: Cellular Heterogeneity–Adjusted cLonal Methylation (CHALM) improves prediction of gene expression.
nature.com
Nature Communications - Here, the authors introduce Cell Heterogeneity–Adjusted cLonal Methylation (CHALM) as a methylation quantification method that considers the heterogeneity of sequenced...
6
12
81
RT @XShirleyLiu: Twenty years of Genome Biology. Quite honored that our MACS is one of the top papers published in the last two decades, to….
genomebiology.biomedcentral.com
Genome Biology is a leading open access journal in biology and biomedicine research, with 9.4 Impact Factor and 14 days to first decision. As the ...
0
28
0
RT @LeiLi_bioinfo: Super Excited to share our new pre-print describing the first atlas of human 3'QTLs, i.e. ~0.4 million genetic variants….
biorxiv.org
Genome-wide association studies have identified thousands of non-coding variants that are statistically associated with human traits and diseases. However, functional interpretation of these variants...
0
14
0
Congratulations Ping.
Dr. Ping Ji has received a new two-year award from @theNCI to study factors in the development and progression of glioblastoma multiforme, the most common and aggressive brain tumor. Dr. Ji is an Assistant Professor in @bmb_utmb. We congratulate him on this research opportunity!
0
1
2
Homeobox oncogene activation by pan-cancer DNA hypermethylation
genomebiology.biomedcentral.com
Background Cancers have long been recognized to be not only genetically but also epigenetically distinct from their tissues of origin. Although genetic alterations underlying oncogene upregulation...
0
8
14