Siegel Lab
@siegel_lab
Followers
1K
Following
2K
Media
57
Statuses
935
We use T. brucei to study the role of chromatin and genome architecture in antigenic variation.
Munich, Germany
Joined April 2018
we have stopped using twitter/X. please follow us on @siegel-lab.bsky.social.
0
0
1
RT @leslievosshall: Imagine if you woke up this morning and found that the NIH-funded database that holds all of the data that you and thou….
0
116
0
RT @BSPparasitology: 🚨 #BSP2025 Spring Meeting: Joint meeting of @BSPparasitology, @SgtpSsmtp, & @DParasitologie! 🚨. - 1st time ever: the B….
0
48
0
RT @parasitesrule: Registration and abstract submission is now open for the #KMCB meeting. Meeting will take place in Woods Hole, MA from S….
parasitesrule.com
Program (updated Sept. 5)
0
18
0
RT @luisamfigueired: Hey Parasitology trainees, join us at the Kinetoplastid Molecular and Cell Biology meeting in September in Woods Hole.….
0
12
0
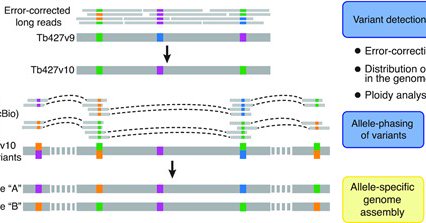
Contact us, If you have any questions about a particular region of the genome or your favorite cell line. Some of these observations are discussed here: Cosentino et al NAR Genom Bioinform 3, lqab082 (2021).
academic.oup.com
Abstract. To date, most reference genomes represent a mosaic consensus sequence in which the homologous chromosomes are collapsed into one sequence. This a
0
0
2
👀 Don't miss out on the super complementary "back-to-back" pre-print from @MonicaMugnier's lab, delving into mosaic VSG formation in vitro and in vivo!📰
biorxiv.org
Antigenic variation, using large genomic repertoires of antigen-encoding genes, allows pathogens to evade host antibody. Many pathogens, including the African trypanosome Trypanosoma brucei, extend...
0
3
12
🌟Kudos to @KirstyRMcW, Zhibek Keneshkanova, and @CosentinoRaul for leading this remarkable endeavor! Dive in and share your thoughts!.
1
0
0
We’re thrilled to share our pre-print, unraveling the determinants of antigen expression hierarchy! @Cell2_Cell @ERC_Research @LMU_BMC A fantastic collaboration with the labs of @colomemaria_epi and @MonicaMugnier.🎉 Check it out ➡️
biorxiv.org
Antigenic variation is an immune evasion strategy used by many different pathogens. It involves the periodic, non-random switch in the expression of different antigens throughout an infection. How...
3
9
36
RT @robert_ife: Publication alert ! Paper from in @Nature .How complex epigenetic modification signatures regulate….
0
62
0
RT @HAKIMIMohamedA1: It's time to thank VEuPathDB and the dedicated people behind the screens who help us run great omics studies!.
0
4
0
Here is the GitHub repo: Great work by @SchmidtMarkusR, Anna Barcons-Simon, and @claudia_rabu! .Thanks to our funders @crc_1064 @ERC_Research @dfg_public.
github.com
On-the-fly processing and visualization of interactome data - Siegel-Lab/BioSmoother
0
0
2
"Smoother: On-the-fly processing of interactome data using prefix sums" has been published in @NAR_Open.🥳Check it out to learn how #HiC and RADICL-seq data can be processed and visualized using an interactive interface. #Genomics #Bioinformatics
3
6
26
RT @JoanaRCF: (1/8) Very excited to share our latest work now published in @NatureComms. The result of a joint effort between Faria & Horn….
0
24
0
Great effort to understand the very complex genome architecture of T. cruzi.
Very happy! Our work was published! @fdiazviraque ML Chiribao, Gaby Libisch congrats! Thanks to @IPMontevideo @fmedoficial @FillingV .#Trypanosoma #chromatin #DNAmethylation #geneexpression .
0
1
7
RT @GEWard14: Nina Papavasiliou and I are pleased to announce the program for the 2024 Host-Parasite Gordon Conference: .
0
42
0