Nathan Sheffield
@shefflab
Followers
43
Following
16
Media
1
Statuses
42
Joined May 2017
I'm hosting a virtual open house today at 2-3pm Eastern Time -- Drop in if you're interested in hearing about getting a PhD in Computational Biology at UVA.
compbio.med.virginia.edu
Join our virtual open house sessions to meet faculty, students, and administrators from the UVA Computational Biology PhD program.
0
0
0
Interested in learning about doing a PhD in computational biology? Join us for open office hours tomorrow at 11am Eastern.
compbio.med.virginia.edu
Join our virtual open house sessions to meet faculty, students, and administrators from the UVA Computational Biology PhD program.
0
0
0
Taming the reference genome jungle: the refget sequence collection standard https://t.co/A3etiUi6zF
#biorxiv_genomic
biorxiv.org
Reference genomes are foundational to genomics but suffer from widespread ambiguity and incompatibility due to inconsistent naming, undocumented differences, and lack of formal mechanisms for...
0
1
6
We launched a new website for our PhD program in computational biology at UVA. Visit:
compbio.med.virginia.edu
UVA's PhD Program in Computational Biology combines AI, data science, and biology to prepare the next generation of scientists. 100% funded positions with world-class faculty.
0
0
0
Need to combine BED files into a consensus set? Our paper with methods to do this just came out in NAR! --Methods for constructing and evaluating consensus genomic interval sets
academic.oup.com
Abstract. The amount of genomic region data continues to increase. Integrating across diverse genomic region sets requires consensus regions, which enable
0
1
4
BEDMS: A metadata standardizer for genomic regionattributes https://t.co/Tk3Vp9o3kv
#biorxiv_genomic
0
2
0
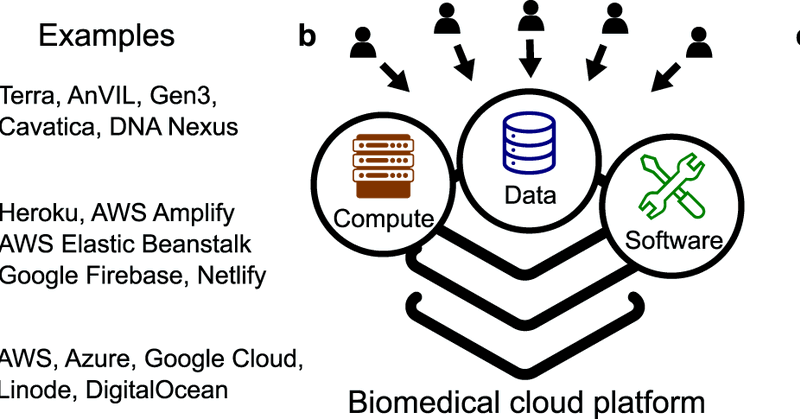
From biomedical cloud platforms to microservices: next steps in FAIR data and analysis: https://t.co/CwjMiKzW7S -- Read our new perspective on biomedical cloud computing at Scientific Data.
nature.com
Scientific Data - The biomedical research community is investing heavily in biomedical cloud platforms. Cloud computing holds great promise for addressing challenges with big data and ensuring...
0
6
11
Our paper on standardizing sample metadata is out in GigaScience:
0
0
5
ATAC-seq pipeline PEPATAC paper now published in NARGAB:
academic.oup.com
Abstract. As chromatin accessibility data from ATAC-seq experiments continues to expand, there is continuing need for standardized analysis pipelines. Here
0
0
3
New bioinformatics paper: Embeddings of genomic region sets capture rich biological associations in lower dimensions
academic.oup.com
AbstractMotivation. Genomic region sets summarize functional genomics data and define locations of interest in the genome such as regulatory regions or tra
0
0
3
Identity and compatibility of reference genome resources paper is out in NARGAB:
academic.oup.com
Abstract. Genome analysis relies on reference data like sequences, feature annotations, and aligner indexes. These data can be found in many versions from
0
1
8
Refget: standardised access to reference sequences https://t.co/Efonf4PiRL
#biorxiv_bioinfo
0
1
4
Identity and compatibility of reference genome resources https://t.co/3v7Cf4gHph
#biorxiv_genomic
0
1
1
Anyone else interested in switching from HPC environment modules to linux containers to manage shared group software? Here's how I did it:
0
0
0
Ref genie is some pretty slick work from @shefflab. If you don’t know, now you know.
@tangming2005 My lazy approach has been to use refgenie when possible: https://t.co/P3tJvpZuhL It's pretty easy to pull things and if someone asks which versions of files you used, you just link the repo.
0
5
5
Bedshift: perturbation of genomic interval sets https://t.co/94fUbWEr6P
#biorxiv_bioinfo
0
1
3
Great example of multiple awesome #communities working well #together. Honored to have been able to contribute. #usegalaxy #refgenie #cvmfs #referencedata #opendata
Refgenie assets can be the source of reference datasets in Galaxy @galaxyproject Happy to share the preprint on the integration of refgenie into the Galaxy platform:
0
4
13
PEPATAC: An optimized ATAC-seq pipeline with serial alignments https://t.co/KPM2tjPJ0d
#biorxiv_genomic
0
1
4
Linking big biomedical datasets to modular analysis with Portable Encapsulated Projects https://t.co/TSscaZuWPY Code: https://t.co/4cVc0RWIrW docs: https://t.co/xtUDLbNC7U. Really useful stuff out of @shefflab
0
6
12