Richard Stöckl
@richard_stoeckl
Followers
176
Following
4K
Media
4
Statuses
1K
PhD Student at the German Archaea Centre @uni_regensburg. #Microbiology #Bioinformatics. @[email protected] @richard-stoeckl.bsky.social
Joined March 2019
Our magnetic beads separate DNA based on its size. You can isolate the fraction, for example, of 200-700 bp, removing shorter and longer DNA strands. The DNA molecules stick to the carboxylated surface, and the strength of sticking depends on the size of the DNA. For some
@pochekailov Tell me more about your beads
16
36
337
skani v0.3.0 is released. https://t.co/dEkIzxIbDr * 30-40% potential reduction in memory * Breaking changes to indexing and searching databases Calculate ANI for contigs, genomes. Search vs > 140k genomes: pre-indexed GTDB-R226 available for download.
github.com
Fast, robust ANI and aligned fraction for (metagenomic) genomes and contigs. - bluenote-1577/skani
0
23
54
Stoked to finally have a preprint out for Phold, our tool that uses protein structural information to enhance phage genome annotation
1
6
28
@McCulloughFund No, it did not. At best, it found that people with cancer or other diseases have different expression profiles. But it absolutely does not show any relationship with mRNA vaccines. It is a severely flawed study. Read my review here: https://t.co/8PekiPMak5
scienceintegritydigest.com
Today, 25 July 2025, a preprint was posted claiming that significant gene expression changes were found in individuals with new-onset cancer and other diseases after receiving mRNA COVID-19 vaccine…
0
11
36
this is crazy (spectrogram of training data vs reproduced) now you think about it, birds probably also feel "this is crazy" when they hear us talking to each other
Bird saves and reproduces data: A PNG image of a bird ( photo of a bird -> spectral synthesizer ) was reproduced by an adult Starling bird https://t.co/eW4G6d4kdn. It seems to have reproduced the sound in conjunction with some additional notes which made it not detectable
16
90
1K
It has been a while… but an updated, faster & more accurate version of OrthoFinder is now out! Scales to thousands of species on conventional compute resources with higher accuracy than ever before and the same data rich fully phylogenetic outputs 👉 https://t.co/VivnkqSyKs
biorxiv.org
Here, we present a major advance of the OrthoFinder method. This extends OrthoFinder’s high accuracy comparative genomic framework to provide substantially enhanced scalability and accuracy. Specif...
1
62
153
For many of those who were asking on BLOW5 vs POD5 for nanopore signal data, here is a finally detailed benchmark we did: https://t.co/ZspXSlrW9e Summary: performance of BLOW5 is >= POD5 (from ~= to 100X, see below), with benefit of having ~3 dependencies instead of >50.
1
19
55
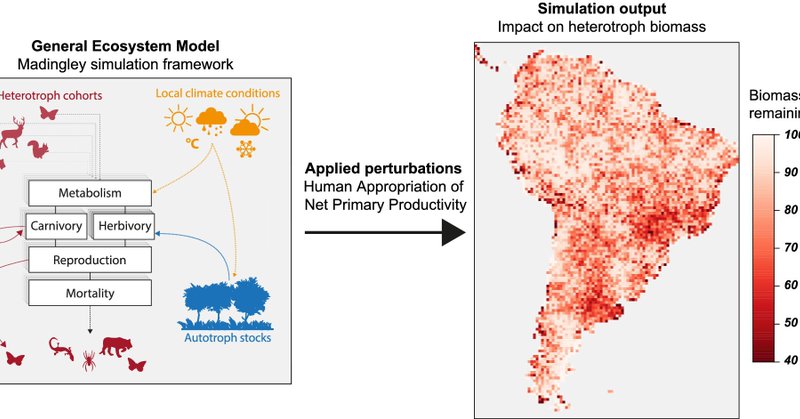
Our new article “Advancing general ecosystem models (GEMs): Towards a mechanistic understanding of the biosphere in the light of the Anthropocene” let by Joachim Paul Töpper just out at Ecological Solutions and Evidence https://t.co/Apl5FrGiAp
besjournals.onlinelibrary.wiley.com
General ecosystem models (GEMs) have been framed as the ‘climate model’ equivalents for nature, but while the latter have been continuously improved and diversified for several decades already, the...
5
8
30
A genomic catalog of Earth's bacterial and archaeal symbionts
biorxiv.org
Microbial symbiosis drives the functional and phylogenomic diversification of life on Earth, yet remains underexplored due to culturing challenges. This study employed machine learning (ML) to...
0
10
33
Functional prediction of proteins from the human gut archaeome #microbiology #microbiota #archaea
@ISMEComms @ISME_microbes
https://t.co/JeKm17pjhr
0
3
10
Very proud to announce that the Windows Subsystem for Linux is now open source! https://t.co/2yJVi2Xc6U
blogs.windows.com
Today we’re very excited to announce the open-source release of the Windows Subsystem for Linux. This is the result of a multiyear effort to prepare for this, and a great closure to the first ever...
138
1K
9K
Expanding the cultivable human archaeome: Methanobrevibacter intestini sp. nov. and strain Methanobrevibacter smithii ‘GRAZ-2’ from human faeces. Available to read in #IJSEM: https://t.co/LZCRxGaDb8
0
5
13
Out now in @MolecularCell: Cyanobacterial Argonautes and Cas4 family nucleases cooperate to interfere with invading DNA https://t.co/6hVLt4JGOG Most long-A pAgos interfere with invading DNA solo. Why then are cyanobacterial pAgos co-encoded with a Cas4-like protein? A 🧵
1
17
46
Published in @MicrobioSoc
https://t.co/Cp0GR0hb74 highlighting: - Contribution of #prophage dynamics in the #pangenome evolution - Pangenome data structure to trace orthologous prophages - Generalist & specialist prophages in the Pectobacterium genus
0
5
7
9th @denbiOffice "Microbial Genomics training course" now open for registration! 3 sessions full of microbial bioinformatics at JLU Giessen: I: QC & QA, assembly II: regional & functional annotation III: comparative genomics Info & registration:
0
3
6
Just casually doing some SUP basecalling on an AMD 7900XTX GPU 😅 with ROCm in a workstation at home. A work in progress by Hasindu and Bonson in the lab.
1
2
13
New paper out! We discovered a tiny archaeon with the smallest known archaeal genome — only 238 kbp! Candidatus Sukunaarchaeum mirabile has almost no recognizable metabolic pathways and may rely heavily on a host to survive. https://t.co/0T5XDRaiyM
2
30
91
I got hit by some rather sudden and extreme financial hardship so if anyone is in need of remote wetlab contract research, strictly BSL1, do let me know. Currently scrambling for gigs. Plant, Bacterial, Archaeal Bioeng Custom Lab Hardware Turn Key Genetic Design Please RT 💚
19
336
531