Laura Chacon Machado
@pomarros4
Followers
49
Following
622
Media
7
Statuses
22
Microbiology PhD candidate at @Cornell @JoePetersLab Interested in bacterial genetics, MGEs are my favorites 🧬✨
Joined May 2023
De novo engineered guide RNA-directed transposition with TnpB-family proteins reveals features of naturally evolved systems
biorxiv.org
Programmable DNA integration using CRISPR-associated transposons (CASTs) offers powerful capabilities for genome engineering. The single effector Cas12k CAST examples evolved from a fixed guide TnpB...
1
9
40
Tn7 family transposons that include all guide RNA-directed transposons (CAST) are common in bacteria. We now find diverse representatives across archaea, many in the Asgard group. Reviving an Asgard element confirmed the exciting behaviors of the family
biorxiv.org
Tn7 family elements are DNA transposons in bacteria that show tight control of targeting, including multiple varieties that evolved guide RNA-directed transposition. We find diverse Tn7 elements...
0
9
59
Our preprint is now published
science.org
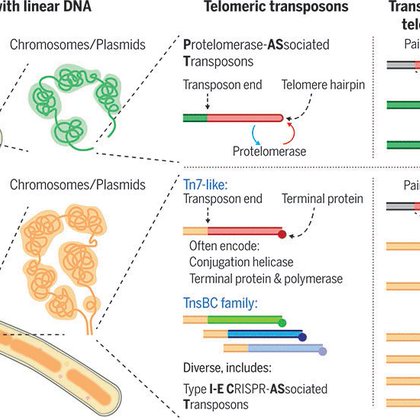
Eukaryotes have linear DNA, and their telomeres are hotspots for transposons, which in some cases took over telomere maintenance. We identified several families of independently evolved telomeric...
Eukaryotes have linear DNA and their telomeres are hotspots for transposons. Many bacteria are also known to have linear genomes. We identified several families of independently evolved telomeric transposons. 1/7
3
7
22
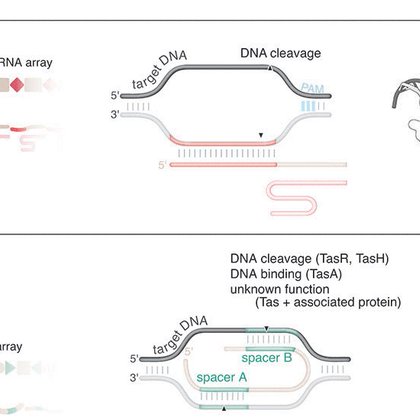
We are excited to report the discovery of TIGRs, a widely-occurring RNA-guided system found in bacteria and their viruses. TIGRs consist of a peculiar repeat region which is transcribed into RNA and processed into multiple guide RNAs to direct TIGR-associated proteins to their
science.org
RNA-guided systems provide remarkable versatility, enabling diverse biological functions. Through iterative structural and sequence homology-based mining starting with a guide RNA-interaction domain...
26
271
1K
CRISPR-Cas systems are common in chromosomes and on mobile plasmids. Therefore, targeting arrays would provide a stable integration position facilitating vertical transmission while maintaining the ability to mobilize between bacteria for horizontal transmission. 10/10
0
0
0
We did not find significant array degradation downstream the insertion. We suspect that integrating into a spacer in the arrays and keeping a small size allow these elements to not disrupt CRISPR-Cas system function. 9/10
1
0
1
Aligning the DNA sequences that flank the ends of the tRNA- and array-targeting elements revealed a conserved DNA motif 40 to 50 bp away from the transposons left ends. 8/10
1
0
0
Unlike the tRNA-targeting pathway from the closely related McCAST element, the array targeting element was not functional in E. coli and may need host factors not found in this heterologous host. 7/10
1
0
0
AlphaFold3-generated structures of TnsD proteins from A. franciscana (AfTnsD, array-targeting) and M. californica (McTnsD, tRNA-targeting) suggest that the C-terminal region absent in AfTnsD corresponds to a helix-turn-helix domain (HTH) used for DNA binding. 6/10
1
0
0
Unlike most Tn7-like elements, array-targeting transposons do not accumulate cargo genes. This contrasts with what is found even with the closely related tRNA-targeting elements. 5/10
1
0
0
A distinct structural difference that co-occurs with the change in att site used between these two elements is a C-terminal motif found in the tRNA-targeting TnsD proteins that is absent in the array-targeting group. 4/10
1
0
0
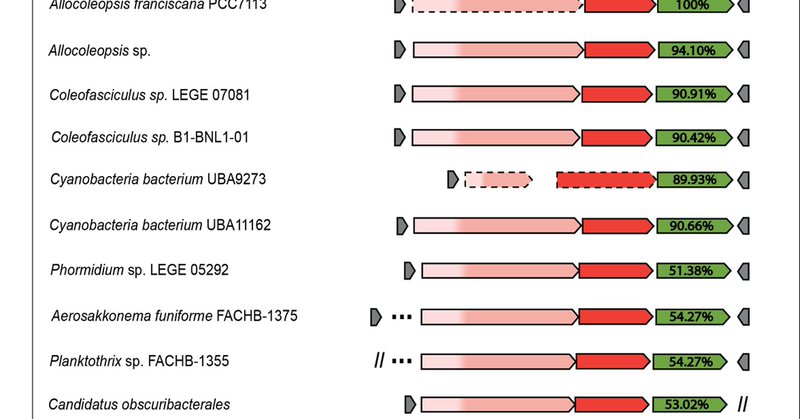
A similarity tree of nonredundant TnsD/TniQ proteins revealed that the array-targeting transposons are phylogenetically close to TnsD-mediated tRNA-targeting transposons in this phylum, a group that includes validated I-D and I-B2 CAST. 3/10
1
0
0
We found a novel family of Tn7-like transposable elements in Cyanobacteria which display a preference for insertion within CRISPR arrays, suggesting a previously unrecognized functional interplay between Tn7-like elements and CRISPR-Cas systems. 2/10
1
0
0
Our preprint describing a family of Tn7-like transposons that evolved to target CRISPR repeats is now published at Mobile DNA #Tn7 #transposons #mobileDNA 1/10 https://t.co/nkMS5cslyY
link.springer.com
Mobile DNA - Tn7 family transposons are mobile genetic elements known for precise target site selection, with some co-opting CRISPR-Cas systems for RNA-guided transposition. We identified a novel...
3
8
21
Recent work from the lab from Laura Chacon @pomarros4 describing a family of Tn7-like transposons that evolved to target CRISPR repeats being presented at #cshlTE24 and now on bioRxiv
biorxiv.org
Tn7 family transposons are mobile genetic elements known for precise target site selection, with some co-opting CRISPR-Cas systems for RNA-guided transposition. We identified a novel group of...
2
25
55
A family of Tn7-like transposons evolved to target CRISPR repeats https://t.co/wUerbORHhH
#biorxiv_molbio
biorxiv.org
Tn7 family transposons are mobile genetic elements known for precise target site selection, with some co-opting CRISPR-Cas systems for RNA-guided transposition. We identified a novel group of...
0
7
17
It was a pleasure working with @PopoMicro on this review of CRISPR-Cas associated transposons (CAST). Hopefully it gives a sense for the diversity of Tn7-like transposons and CRISPR systems that independently combined. https://t.co/Aq8lz2HG3O
2
40
117
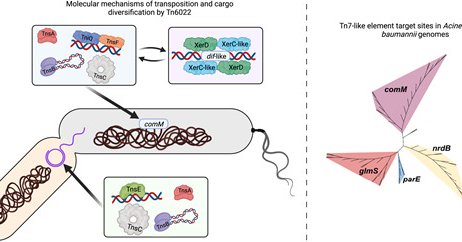
This story involved many hands over many years led by @BertCorrea3 and @SaadleeS. Novel mechanisms of diversity generation in Acinetobacter baumannii resistance islands driven by Tn7-like elements
academic.oup.com
Abstract. Mobile genetic elements play an important role in the acquisition of antibiotic and biocide resistance, especially through the formation of resis
0
15
48