Marc Jan Bonder
@mjbonder
Followers
290
Following
407
Media
6
Statuses
111
Postdoctoral-fellow / team-leader studying the genetics, epigenetics and environmental effects on biological #omics. #DKFZ & #EMBL (former #EBI & #UMCG)
Heidelberg, Germany
Joined December 2015
#vacature We zoeken een #ICT teamleider voor de @MOLGENIS groep @UMCG. Samen ontwikkelen we slimme software voor gezondheids onderzoek. Kun jij zorgen voor team organisatie en persoonlijke begeleiding? Dan doen de (project)leiders de planning. Zie https://t.co/fCvDnnCcfo.
lnkd.in
This link will take you to a page that’s not on LinkedIn
0
3
1
Cultural program of our annual #geneforum23, join us next year when Tartu takes on the role as European Capital of Culture @Tartu2024 #DuoRuut
0
2
22
I am thrilled to share our study on the de novo assembly and characterisation of 43 diverse human Y chromosomes, published today in @Nature, together with the first #T2T Y assembly from @aphillippy, @ArangRhie and co-authors
12
49
158
We are very proud that Monique van der Wijst @MoniquevdWijst has been awarded a VIDI grant for her project to study gene expression variation. Monique, many congratulations! @umcg @univgroningen @NWONieuws
1
4
16
Parallel to this we are working on single-cell eQTLGen as well, a federated single-cell eQTL meta-analysis, where we aim to identify cell-type specific eQTLs. Please have a look at https://t.co/pvj5sdfZke for more information on this exciting project as well.
0
4
4
Are you interested in trans-eQTLs? We are working on a new genome-wide eQTL meta-analysis in blood samples ( https://t.co/pvj5sdfZke). Over 30 cohorts are already contributing to this initiative. Using a federated analysis model, cohorts do not have to share privacy sensitive data
2
38
96
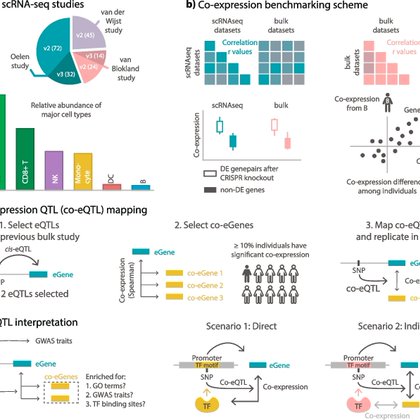
Genetic variation can affect co-expression relationships, as we see in a meta-analysis of single-cell eQTL data, now out: https://t.co/NPDV5DUmKE. We observe colocalisation between a rheumatoid arthritis locus and effects on co-expression of T cell activation genes with RPS26.
genomebiology.biomedcentral.com
Background Expression quantitative trait loci (eQTL) studies show how genetic variants affect downstream gene expression. Single-cell data allows reconstruction of personalized co-expression networks...
2
19
50
Curious about how genetics regulates genes in the brain, and whether this can be linked to disease? I am looking for a PhD student to research just that!
0
12
19
New EMBL research shows that long-read genomic sequencing seems to detect some complex DNA mutations better than short-read genomic sequencing. This provides a tool to better identify, understand, and detect complex DNA mutations that lead to cancers. https://t.co/UQra41smBt
0
7
34
Exciting opportunity to join us as scientific coordinator to shape one or multiple of the collaborative efforts our lab coordinates. Suitable for scientists transitioning into a coordinator role. Data science or more biological background both welcome.
1
4
9
New work from the lab in which we developed a computational method for predicting the age of individual cells from single-cell methylation data https://t.co/eDGmjBYcrQ Fantastic collaboration with @vonMeyenn @mjbonder @OliverStegle @rulandsgroup
2
13
68
Do all cells age the same? At least not by DNA methylation age: Single cell DNA methylation ageing in mouse blood https://t.co/kwuASAnFK3
#bioRxiv. Wonderful collaboration between @mjbonder @ReikLab @OliverStegle @vonmeyenn_lab & @rulandsgroup!
5
37
162
Come work with us! It's an exciting time to work in any one of these fields, let alone all three.
We are HIRING! 🔊 Come join our Computational Biology team at @GSK to work on scientific research and method development for drug discovery using protein informatics, human genetics, and machine learning. Applications open until January 8th:
1
3
10
Happy to share our preprint on the de novo assembly and characterisation of 43 diverse human Y chromosomes, revealing remarkable complexity and diversity in size and structure. #HGSVC
biorxiv.org
The prevalence of highly repetitive sequences within the human Y chromosome has led to its incomplete assembly and systematic omission from genomic analyses. Here, we present long-read de novo...
4
28
80
The toolbox encompass a efficiënt file-format, a segmentation method and differential methylation testing and reporting! Work by Rene Snajder, together with @AdrienLeger2 and @OliverStegle and input of many collaborators. Thanks all!
1
2
4
New work in the ONT methylation space! We had been working on it for a while but want to spread it wider now! We present Pycometh a toolbox for methylation analyse on ONT sequencing data!
biorxiv.org
Advances in base and methylation calling of Oxford Nanopore Technologies (ONT) sequencing data have opened up the possibility for joint profiling of genomic and epigenetic variation on the same long...
2
8
39
Exciting opportunity to join us as scientific coordinator in data science. The role will take key responsibilities to coordinate data science activities at @DKFZ and in the Heidelberg area.
1
17
29
Thanks to @tobias_757 @ReneSnajder @AdrienLeger2 Milena Simovic @OliverStegle @ewanbirney Aurelie Ernst @JanKorbel5 for the great collaboration! @DKFZ @emblebi @embl
1
0
1
We believe our work provides a basis of the broader application of ONT seq in cancer sequencing studies. Profiling complex genomic events as well as the DNA methylation landscape. (3/4)
1
0
1
We: 1) describe a novel type of complex genomic rearrangement, templated insertion threads; 2) Provide a haplotype-resolved assembly of a chromothripsis event; 3) Functionally annotate these events using methylation called from ONT and RNA seq data (2/n)
1
0
0