Adrien Leger
@AdrienLeger2
Followers
867
Following
3K
Media
193
Statuses
2K
Director of Modified Bases Research at @nanopore. Moving slowly to where the sky is less depressing. https://t.co/U30YEgDbsO
Oxford, England
Joined September 2014
🚨 We’re hiring ! 🚨.Join our cutting-edge research team as a molecular biologist at @nanopore HQ in Oxford. Perfect for a fresh PhD or MSc with a couple of years’ experience. Work at the interface of chemistry, molecular/synthetic biology and AI. 👉
0
8
14
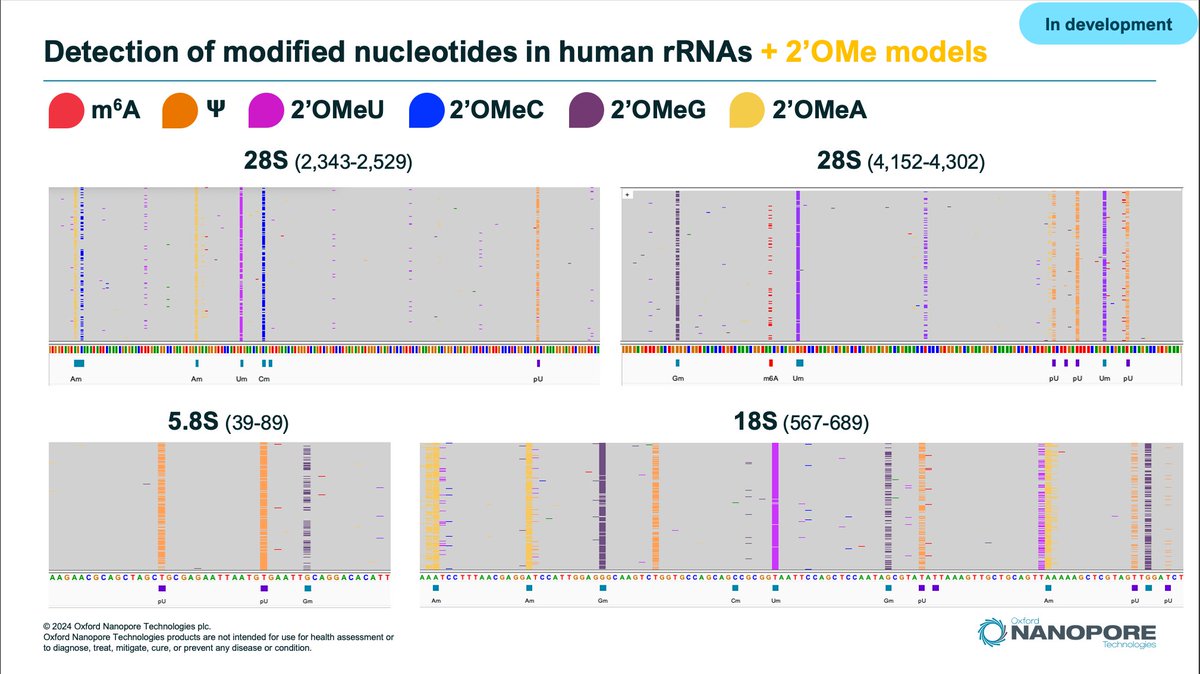
RT @Stoibs11: More details on new RNA modified bases detectable with @nanopore sequencing. #NanoporeConf
0
4
0
RT @Stoibs11: Dorado 1.0 🚀release with new RNA modified base models for 2’Ome as well as improvement of all DNA and RNA modified base model….
0
13
0
RT @Daniela_Bezdan: Tweet 1/2.WOW—2’O-methylation detection in direct RNA-seq is here! A breakthrough for RNA therapeutics, viral evasion,….
0
4
0
RT @bioinformer: Who has DNA methylation on ~5,000 microbes sequenced w/@nanopore? We do! . Who’s working hard on getting this data out bef….
0
11
0
RT @iiSeymour: Dorado is 1.0! 🎉. This London Calling release updates our DNA and RNA models to v5.2 with a new suite of mod models that are….
0
27
0
RT @nathanbaggers: Excited about @nanopore's collaboration with @uk_biobank to create the first comprehensive epigenetic map of the human g….
0
9
0
RT @Libby_Snell_: I am absolutely thrilled to be back in Kanazawa, Japan (a fave!), for the #tRNA2024 conference! It was my pleasure to pre….
0
1
0
It's Christmas in advance with a very special @nanopore #opendata release ! 🎅.Today we are releasing our 5mC 5hmC and 6mA DNA mod synthetic control datasets with each mods in all possible 5 mers contexts. All nicely wrapped in a comprehensive validation blog post.🎁.
New blog: Modified Base Best Practices & Benchmarking for @nanopore 🧬 sequencing Includes:. * Raw POD5 files for synthetic ground truth strands.* Modified base calling best practices.* Modkit recommendations. Check it out: #methylation #bioinformatics.
0
13
35
RT @Stoibs11: New blog: Modified Base Best Practices & Benchmarking for @nanopore 🧬 sequencing Includes:. * Raw POD5 files for synthetic gr….
epi2me.nanoporetech.com
Modified bases, including methylation, regulate many biological processes - from eukaryotic gene…
0
22
0
RT @slavov_n: The epitranscriptome formed by the growing number of modifications occurring within mRNA transcripts.
0
244
0
RT @RosemaryDokos: Low coverage (5x) @nanopore data vs methylation arrays delivers far more data. Great talk by the wasatch team at #ashg20….
0
6
0
RT @ewanbirney: Some perspective here - like protein modification - phosphorylation, acteylationn etc - and DNA modification - 5mC, 5hmC an….
0
1
0
RT @ewanbirney: When we look at the kmers around the sites, we see enrichment in the DRACH motif and the known m5C motifs, along with novel….
0
1
0