Michael Chiang
@michaelchchiang
Followers
43
Following
23
Media
12
Statuses
34
Biophysics postdoc at Davide Marenduzzo's lab @EdinburghUni @PhysAstroEd
Joined December 2021
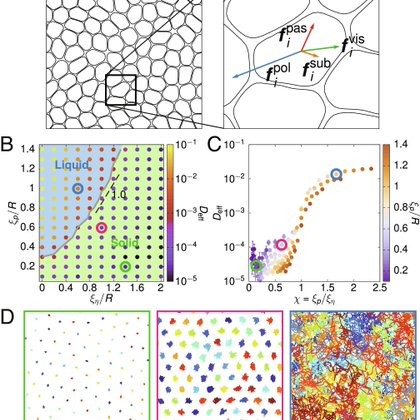
Super excited to share our latest work looking at mechanisms that contribute to local hexatic and nematic ordering in tissue monolayers! Intercellular friction and motility drive orientational order in cell monolayers | PNAS
pnas.org
Spatiotemporal patterns in multicellular systems are important to understanding tissue dynamics, for instance, during embryonic development and dis...
0
1
3
We are excited to announce our new manuscript is on bioRxiv https://t.co/ZFDu21NZRJ. We use super-resolution imaging, molecular biology and polymer simulations to show that nuclear RNA forms an interconnected network of microgels. Well done everyone!
4
53
219
M. Chiang, A. Hopkins, B. Loewe, M. Cristina Marchetti, D. Marenduzzo, Intercellular Friction and Motility Drive Orientational Order in Cell Monolayers. arXiv [cond-mat.soft] (2023).
0
3
6
@IanMurrayMP Could you please RT this post to further raise awareness of the incident and increase the likelihood of finding a witness? It happened not far from your office. 🙏
0
0
0
Could everyone in Edinburgh please RT this to maximise the chance of finding a witness of the incident? I suffered a head injury and broke a few teeth because of it and so far there has been no luck with any CCTV footage. The exact timing was more like 8:15am on that day
We are appealing following a crash on Ratcliffe Ter, 8.45am Thurs 26 Jan, involving a blue Vauxhall Corsa and a cyclist. A man aged 27, the cyclist, was taken to ERI for treatment. If you saw what happened or have dash-cam please call us on 101 - Inc 0654 of 26/1/23
15
290
126
So far, most DNA-loop-extrusion data were taken on naked DNA. But what happens on chromatin in cells where DNA is loaded with many proteins? Will SMC proteins (that are fast but weak motor proteins) get blocked? Or find a way to translocate the roadblock into the extruded loop? /
4
51
206
We have a new postdoc position available in our collaborative research group. Using molecular biology and imaging techniques to study and understand transcriptional noise. Get in touch if you would like to chat or apply here https://t.co/qjZSMdy7cb
1
16
22
It's been an awesome week learning and discussing new science at the #EMBOworkshop on Nuclear Structure and Dynamics. Many thanks again to the organisers for giving me the opportunity to speak about our lab's work on HiP-HoP and 3DGene ( https://t.co/JvT6kcFfiG)!
0
2
15
What a fantastic experience so far at the Genome Organisation by SMC Complexes conference! It's been great to present my simulation work on 3D gene structure and transcriptional noise and learn about all the exciting new science of SMC proteins. #BioChemEvent
0
1
13
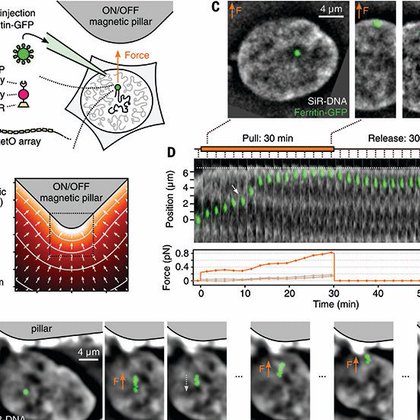
Our work on chromosome micro-manipulation in living cells is out in @ScienceMagazine! https://t.co/BCzYOuedcm What if you could put your hand 🫳 inside a nucleus, grab a small portion of a chromosome 👌, and see how it reacts when you pull on it? 🤔 … 🧵 1/n
science.org
An approach for probing the force response of interphase chromosomes in living cells reveals their material nature.
23
171
722
One new application of the DFCC method for studying nuclear organization. Check out our new story on how RNA Pol II movement during transcription is coordinated over long-range in the context of chromatin dynamics, https://t.co/xqFvCASWUV
0
12
59
A cool web tool for browsing the simulated gene structures from our paper 'Gene structure heterogeneity drives transcription noise within human chromosomes'. https://t.co/teVJ9IEvOd Here is one of my favourite genes - what's yours?
Check our new website ‘3DGene’. With @PhysAstroEd we have modelled the 3D structure of all genes in the human genome and predicted transcription. https://t.co/X36ACQnKrj There are a few glitches with rendering so adjust bead size to uniform for best results.
0
4
6
Overall, our work showed two major links between gene structure and transcription: (1) the formation of phase-separated TF condensates drives average transcriptional activity. (2) diversity in gene topology and loop extrusion contribute to transcriptional noise. 10/10
0
1
2
To validate the correlation between noise and loop extrusion, we ran simulations without extrusion. In line with previous work, we saw no notable change in the average transcriptional activity of the genes, but there was a significant decrease in their transcriptional noise. 9/10
1
0
0
Our predicted transcriptional noise correlates significantly with structural diversity. Surprisingly, noise also correlates with the distance between the promoter and the nearest CTCF site and the frequency of loop extruders passing by the promoter. 8/10
1
0
0
The variance (width) of the distribution of TF binding frequency provided us with a way to predict the transcriptional noise of each gene. We found that this correlates with the variability in transcript counts from single-cell RNA-seq data. 7/10
1
0
1
We showed that TFs in the simulations micro-phase separate due to the bridging-induced attraction. We found genes that are close to a TF condensate have higher predicted transcriptional activity. 6/10
1
0
0
We found HiP-HoP can predict the transcriptional activity of each gene. We computed the frequency of TF/protein binding at each promoter, obtaining a distribution of this measure from different simulation runs. The mean TF binding frequency correlates with GRO-seq signal. 5/10
1
0
0
We counted how many ways (or topologies) these partners interact with the promoter. Across all genes, there is large variability in their number of topologies and how the structures are distributed among the topologies, as quantified by a diversity/entropy score. 4/10
1
0
1
We studied how promoters interact with other regulatory elements (enhancers; identified by ATAC peaks). We determined ATAC partners for each promoter and called highly interacting ones as influential nodes. Genes with more of these nodes have more tissue-specific GO terms. 3/10
1
0
1