Mary Makarious
@mbmakarious
Followers
388
Following
4K
Media
31
Statuses
419
egyptian/مصرية 🇪🇬𓂀 • @DataTecnica • currently @NIH studying neurodegen. *omics 🧬🧠 • @geno_ml core dev • @LoyolaChicago @UCLIoN alumna • she/her
washington dc-ish
Joined August 2012
GP2 release 7 is now live!!! Massive effort from great collaborators across @DataTecnica , @NIHAging , @Verily , MJFF and @ASAP_Research to get this done. Check out the release that is now live on Verily Workbench and Terra.
1
11
29
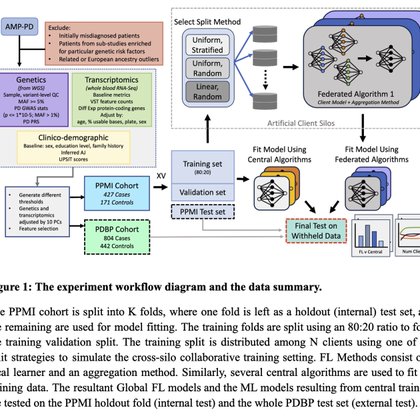
Paper out! Federated learning for multi-omics: A performance evaluation in Parkinson’s disease TL/DR: Federated learning particularly in a boosted framework is comparable to central analyses ... let's start learning across data silos now! https://t.co/A0dKkSFHTU
1
6
20
M&M at it again - honored to present this morning in the beautiful Cartagena about future directions, analyses, and data for @ASAP_Research's GP2!
Lucky to team with the amazing @mbmakarious to talk data engineering for accelerating #openscience at @ASAP_Research GP regional meet in Cartagena. Sidenote, the dual monitors are quite bright. 🫥 📸 @SingletonNeuro
0
0
5
Get some rest and use this as the reason (after reading this) 😴
Get some sleep! Sleep issues in 2M samples show risk of neurodegeneration 15 years out and compensates for low genetic risk. Check the link for a break down of our preprint from a collaboration between the UKDRI, NIH's CARD and @DataTecnica. https://t.co/I3vOfRhGsy
0
0
8
New blog post on our proof of concept for federated learning applications in healthcare. https://t.co/uVj2RSaOkJ We show in a multi-modal precision medicine application, federated machine learning is comparable to centralized data efforts but is safe and efficient across silos!
datatecnica.com
Data silos are a huge issue in machine learning, particularly in the healthcare and biotech settings. This can be due to distributed servers or cloud localities, policy barriers like GDPR, or people...
1
2
5
Hats off to this trio of outstanding scientists for their huge contributions to the field of PD genetics. Congrats to my mentor @SingletonNeuro - Thanks for your tireless effort, perseverance and a never-give-up attitude. What a daily source of inspiration!
Congratulations to Thomas Gasser, Ellen Sidransky and Andrew Singleton, who win the 2024 Breakthrough Prize in Life Sciences for discovering key genetic risk factors for Parkinson's disease. https://t.co/uLhtm3bmHk
@NIHAging @NIH @genome_gov @DZNE_en @uni_tue
0
3
11
Proud of our guy @SingletonNeuro !!!
Congratulations to Thomas Gasser, Ellen Sidransky and Andrew Singleton, who win the 2024 Breakthrough Prize in Life Sciences for discovering key genetic risk factors for Parkinson's disease. https://t.co/uLhtm3bmHk
@NIHAging @NIH @genome_gov @DZNE_en @uni_tue
2
6
30
So honored to be part of GP2 and be a part of this work! 😊
Check out @ELeap29 blog on #GBA1Discovery and the way #GP2research is pioneering collaborative science
2
2
10
(1/2) Thanks to @NewsHour for giving me the opportunity to chat with @johnyangtv about the groundbreaking #GBA1discovery from @ASAP_Research's #GP2 researchers! Intentional #collaboration + #inclusive research paves the way for #discoveries 🙌🏾🙌🏾 We're moving full speed ahead!
New research has identified a genetic variant that increases the risk of Parkinson’s in people of African descent, and is not seen in those with European ancestry. @johnyangtv speaks to @ELeap29 for more.
1
9
35
👏👏👏
Today I presented my project on #GP2AIM. It is a pleasure to talk about the ancestry in association studies and the positive impact in addressing this information in the studies. Let's make science more inclusive
0
0
8
Link to the manuscript can be found here:
academic.oup.com
Makarious et al. investigate the contribution of rare genetic variants, with minor allele frequencies <1%, to Parkinson’s disease risk. They confirm the
1
1
3
However, further research into the biological mechanisms is critical to confirm the role of these genes in PD. Further replication in larger datasets that prioritize familial PD cases and non-European ancestry will provide greater insight into all nominated genes
1
0
2
Additionally, in this work, we have identified mutations in B3GNT3 and TREML1 to be potentially associated with increased risk of PD as well as previously linked with neuroinflammation
1
0
1
Studying just coding variants in this way has reaffirmed mutations in genes previously associated with PD (LRRK2/GBA1) and that while it's likely it's because we're underpowered, it's even more likely that non-coding or SVs are driving the hits we see on a GWAS
1
0
1
(a neat bonus, OpenTargets has added these results to their platform!) https://t.co/7pUSOZ8hqw
Have a look at the recent @mbmakarious et al. rare variant burden testing in Parkinson’s disease, in the context of other genetic information. This and much more are in the latest @OpenTargets Platform release
1
0
2
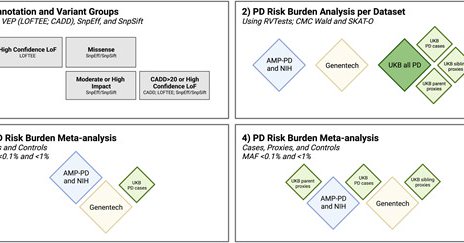
We analyzed rare and ultra-rare variants collapsed by gene across 4 variant classes: Missense, predicted to have moderate or high impact, high-confidence LoF, and variants that are high-confidence LoF or have a CADD score > 20 across the UKBiobank, AMP-PD, NIH, and Genentech
1
0
1
Out now in @Brain1878! 🥳 Here, we conduct the first large-scale study to investigate rare variant associations of Parkinson's disease risk at a genome-wide scale. In total, we looked at 7,184 PD cases, 6,701 proxy cases, and 51,650 healthy controls
New research from @mbmakarious @cornelisblauw & colleagues presents the largest #Parkinsons genetic burden test to date; identifying GBA1 & LRRK2 as 2 genes harboring rare variants associated with PD & nominating several other previously unidentified gene https://t.co/eRI2za32ln
4
13
29
Our own @C_WSolsberg @halsty and friends from NIH CARD 🧬 identify novel AD loci by leveraging diversity across populations @UCSFmac
And another one … @C_WSolsberg and @HamptonLLeonard cranking out #AlzheimersDisease multi-ancestry meta-analysis #GWAS. With some deep dives into heterogeneity of effects and risk prediction implications. Team effort from NIH’s CARD and friends! https://t.co/zPvCbN6SXO
1
6
25
Been a great day for publications from the team. @mkoretsky1 and the team great job on showing genetic overlap and in some instances a depletion of genetic risk across and within neurodegenerative diseases.
Genetic risk factor clustering within and across neurodegenerative conditions (such as #Parkinsons, #Alzheimers, #ALS) - from @HamptonLLeonard & colleagues; "Neurodegenerative diseases have more overlapping genetic etiology than previously expected" https://t.co/BXdQ4FUCKi
1
6
13
Mike is right, Chelsea Alvarado (twitterless 🥲) and @HamptonLLeonard's exciting work here is just the beginning... 🙏
New preprint from the team at CARD, nominating novel drug targets for neurodegenerative disease using multi-omic and genetic data. Hoping this is a starting point for community target discovery specifically focused on ADRD. https://t.co/grFO1wSgP8
0
0
1