Florian Leese
@leeselab
Followers
3K
Following
7K
Media
457
Statuses
3K
Biologist, professor | Studies aquatic biodiversity in the anthropocene w great team | Genetic methods, multiple stressors | not active here anymore
Essen, Nordrhein-Westfalen
Joined March 2015
I will not be active here anymore 📉. All updates on our group activities; thoughts on genetic biodiversity research on #BlueskySocial or #LinkedIn
@leeselab
0
0
1
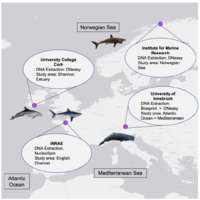
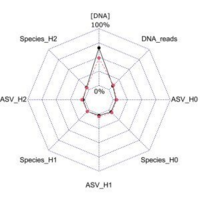
Inter-laboratory ring test for environmental DNA extraction protocols: implications for marine megafauna detection using three novel qPCR assays #eDNA #environmentalDNA
https://t.co/3GLGgnFeZG
mbmg.pensoft.net
The comparability of methods applied to environmental DNA (eDNA) samples across laboratories remains a significant challenge for biodiversity monitoring on a global scale. Performance differences...
0
2
2
🧬 DNA metabarcoding of diatoms is ready for biomonitoring, but varying protocols hinder comparability. @DNAquaIMG (#BiodivMon) shows successful protocol transfer across labs, proposing a strategy ensuring distinct protocols meet minimum standards. https://t.co/U5XgUutftV
mbmg.pensoft.net
DNA metabarcoding of benthic diatoms has been successfully applied for biomonitoring at the national scale and can now be considered technically ready for routine application. However, protocols and...
0
1
12
Congratulations to Dr Katy Klymus, winner of the Metabarcoding and Metagenomics Editors' Choice Award 2024! https://t.co/spzjC3GY0v
mbmg.pensoft.net
Metabarcoding and Metagenomics continues its annual tradition of recognising the year's most outstanding article, as chosen by the journal's editorial team.Congratulations to Dr Katy Klymus et al....
1
3
5
🦈 Do different labs get the same #eDNA results? @ewhale_dna (#BiodivProtect) found differences in detection success, with sharks less often detected. This highlights the need for standardized protocols for reliable biodiversity assessments. https://t.co/BqXj5ac6VC
mbmg.pensoft.net
The comparability of methods applied to environmental DNA (eDNA) samples across laboratories remains a significant challenge for biodiversity monitoring on a global scale. Performance differences...
0
9
13
📣Call for session proposals - #LivingData2025 🐦 Living Data 2025 is a joint conference jointly hosted by @GBIF, @tdwg, @obisnetwork and @GEOBON_org at the @inst_humboldt from 21-24 October in #Colombia. Learn more here: 🔗 https://t.co/sBiKe7TsiE
#Biodiversity #DatosVivos2025
0
10
14
Together with @SeDNASociety, we're committed to promoting the development of #eDNA research. This is why we signed a Memorandum of Understanding with them. We will partner through our @MBMGJournal, including special eDNA issues and collections, and a discount for SeDNA members.
1
2
3
🚨new paper alert! 🚨Inter-laboratory #RingTest for #eDNA extraction protocols: implications for #cetacean and #shark detection using three novel #qPCR assays https://t.co/P3z7AwpVam via @MBMGjournal @MarineBio_LKRod @ewhale_dna
mbmg.pensoft.net
The comparability of methods applied to environmental DNA (eDNA) samples across laboratories remains a significant challenge for biodiversity monitoring on a global scale. Performance differences...
1
8
17
Struggling with inhibitors in your DNA extract? 🚫🧬 Our latest protocol (developed at @leeselab) safely removes inhibitors, ensuring successful amplification every time! Check it out here @protocolsIO : https://t.co/tVXTRggQp7 🔬✨ #DNAExtraction #lowbudget
0
16
72
In this study, researchers find that homogenisation of insect bulk samples yields more comprehensive and cost-efficient biodiversity data than non-destructive lysis. 🔗 https://t.co/9GVsvdV4zZ
#metabarcoding
1
3
4
If you're in the eDNA/metabarcoding/FAIR data practices world, or have interest in it, check out our new paper! #eDNA #FAIRdata #MarineScience The MIEM guidelines: Minimum information for reporting of environmental metabarcoding data https://t.co/ZgVLON5IUZ via @MBMGjournal
mbmg.pensoft.net
Environmental DNA (eDNA) and RNA (eRNA) metabarcoding has become a popular tool for assessing biodiversity from environmental samples, but inconsistent documentation of methods, data and metadata...
0
9
12
The MIEM guidelines: Minimum information for reporting of environmental metabarcoding data #eDNA #environmentalDNA
https://t.co/zMUy5Tp4XA
mbmg.pensoft.net
Environmental DNA (eDNA) and RNA (eRNA) metabarcoding has become a popular tool for assessing biodiversity from environmental samples, but inconsistent documentation of methods, data and metadata...
0
6
11
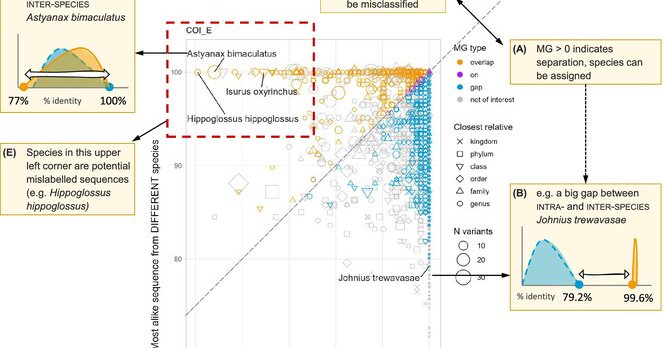
Using Gap Visualization to Navigate Multivariate Metabarcode Data, Select Primer Pairs, and Enhance Reference Data Quality #eDNA #environmentalDNA
https://t.co/5mVRd0917E
onlinelibrary.wiley.com
We introduce the metabarcoding gap visualization and demonstrated how it can make comparisons of PCR primers rapidly comprehensible for researchers in the molecular ecology community. We use gap...
0
1
2
This study of epiphytic diatoms in Iraq shows that a combination of metabarcoding and microscopy can increase the detection and identification of diatom species: https://t.co/4frWBca2wU
#metabarcoding #diatoms #biodiversity @FU_Berlin @uniofgothenburg
0
2
5
We're hiring two bioinformatics-focused PhD students - one on microbial & epigenetic stress response in rivers, another on virus-host stress response in rivers! 🧬🦠💻 @RESIST_CRC
@zwu_ude
@unidue Please RT & spread the word - the @ProbstLab awaits you! 🦸
2
5
9
Gute Neuigkeiten für die @ruhrunibochum @TU_Dortmund @unidue zum Jahresende: der Landtag NRW @landnrw hat die Finanzierung der #ResearchAllianceRuhr gesichert. Wir freuen uns darüber sehr😊Mehr: https://t.co/bvUvY9Eu5U
0
2
7
🎉 First PhD paper of@wolany_lisa comes with good news for #metabarcoding-based #biodiversity monitoring: Homogenized vs „mild lysed“ #insect samples provide highly similar results. We prefer homogenization as it is better scalable, cheaper & identifies slightly more species.
Can insect bulk samples be used in #metabarcoding without homogenising them? Our research shows: it’s technically possible but yields poorer data and is more expensive and time-consuming: https://t.co/y7wxBL2GRx
@leeselab @buchner_dominik @Julian_Enss @MBMGjournal
0
2
19
Interessant: DNA #Metabarcoding liefert Hinweise auf reihenweise unplausible DNA Spuren in #Honig. Vieles spricht dafür, dass nicht drinnen ist was drauf steht. Die Analysen der zwei Labore sehen valide/reliabel aus. Hier sollte genauer hingeschaut werden https://t.co/UjOwC838Gy
0
0
3
What an incredible journey bringing together researchers from across the globe to learn about eDNA metabarcoding from these stellar instructors @leeselab @buchner_dominik @TillMacher & Peter Shum!🗺️🧬 Big thanks to @eDNAcollab for making it all happen! 💡💫
0
4
10