Kristen Curry
@kristendcurry
Followers
72
Following
19
Media
0
Statuses
16
Gut microbiome researcher. Concussion treatment dreamer. PhD student - Treangen Lab, Rice University
Joined October 2020
Thrilled to announce a microbial genomics and metagenomics workshop (CINEMA) that I've organized with @lauren_stadler taking place this summer (July 3rd-10th) at the @RiceUniversity Global Paris Center. Registration page now live (April 15th deadline): https://t.co/b8YK4PYGaC 1/n
events.rice.edu
A metagenome analysis workshop designed for graduate students, postdoctoral fellows, and investigators from diverse backgrounds and biological inte...
1
8
16
The @RiceCompSci team of @traingene and graduate student @kristendcurry have develop Emu, a program that could simplify sorting harmful from helpful bacteria in microbiomes like those in the gut or in agriculture and the environment. https://t.co/t8UD43meFO
0
5
27
Emu, a new microbial community profiling software, identifies bacterial species by leveraging long DNA sequences that span the entire length of a gene. This research, led by Rice CS' @kristendcurry & her advisor @traingene, is out now in @naturemethods. https://t.co/sBpaGts3vR
0
4
9
Excellent to see Emu, led by my @RiceUniversity @RiceCompSci PhD student Kristen Curry (@kristendcurry) & closely developed alongside @AlexDilthey (@HHU_de), among many other collaborators, now out in @naturemethods ! paper: https://t.co/fM7SJrSNM4 code:
gitlab.com
GitLab.com
Emu software uses common gene to profile microbial communities @RiceUniversity @naturemethods
2
6
20
Out now in @naturemethods: Emu, a new method for 16S rRNA @nanopore-based profiling of microbial communities! Great transatlantic collaboration with @kristendcurry, @traingene, et al.. Brief thread follows.
2
35
114
Please join us for a virtual symposium organized by @ricekenkennedy @RiceCompSci, featuring a stellar speaker lineup & taking place on Friday, March 11th (2-year anniversary of WHO declaration of #COVID19 pandemic). Register today! (free but limited spots) https://t.co/AKme2HGEuC
3
28
43
Delighted to share this microbiome + machine learning review article from my group titled: "It takes guts to learn: machine learning techniques for disease detection from the gut microbiome" Superb job by @kristendcurry & @michaelnute @RiceCompSci #ETLS
https://t.co/fH1ban9Q3k
0
6
13
Excited to share work from my first year as a PhD student! Huge thank you to the many collaborators on this project :)
Emu is open-source and readily available for download and use at https://t.co/QHXvzzsVce and also available via ‘conda install emu’: https://t.co/RUvpaOIUPg.
1
3
14
Last but not least, huge thanks to all the co-authors! Tor Savidge lab (Tor & Qinglong), Villapol lab (@svillapol & Sirena), Mendling lab (Werner, Partick, & Enid), Dilthey lab (@AlexDilthey & Alona), & my group @RiceCompSci: @michaelnute, @QiMariaWang1, Libby, and @kristendcurry
0
3
8
Emu is open-source and readily available for download and use at https://t.co/QHXvzzsVce and also available via ‘conda install emu’: https://t.co/RUvpaOIUPg.
gitlab.com
GitLab.com
1
5
6
We also demonstrate the benefits of the EM algorithm in Emu by highlighting the decrease in relative abundance estimation error throughout EM iterations:
1
2
2
Two key questions we focused on for Emu were: 1) Can we improve on relative abundance estimation compared to previous approaches on @nanopore data? 2) Does full-length 16S rRNA analysis w/ Emu offer benefits over Illumina 16S rRNA analyses that are not full length, e.g. V3-V4?
1
2
3
Thrilled to share a new software tool from my PhD student @kristendcurry. Emu is designed for microbial community profiling and leverages an expectation-maximization (EM) algorithm to generate taxonomic abundance profiles from full-length 16S rRNA reads.
biorxiv.org
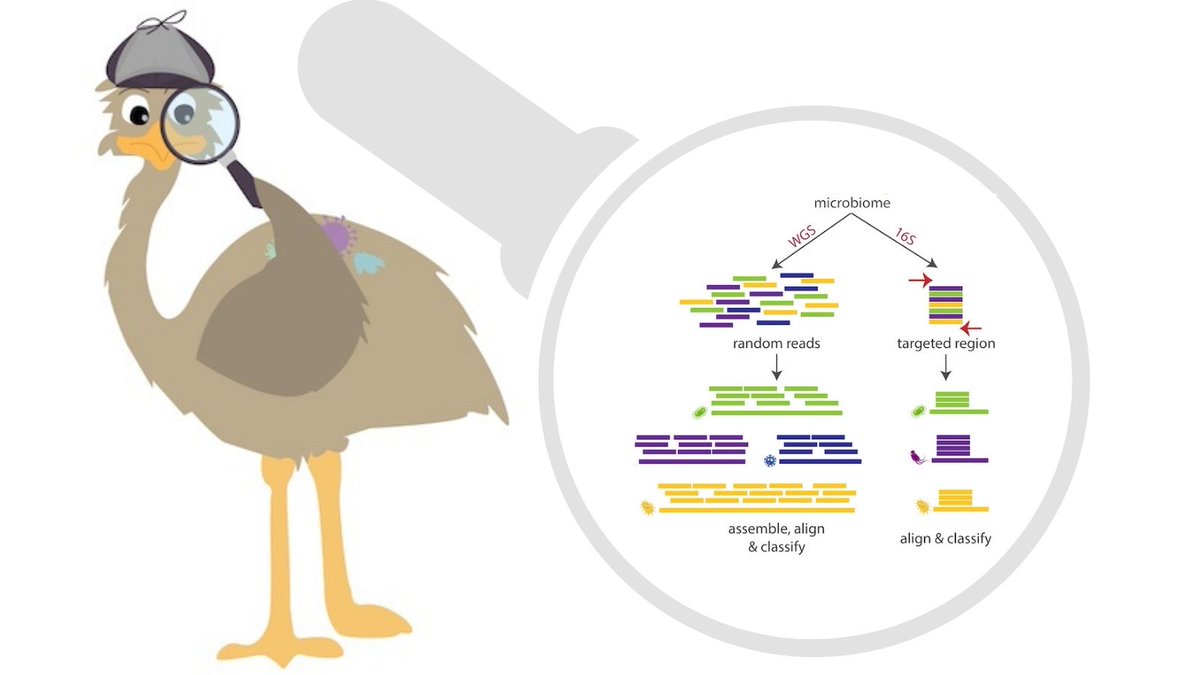
16S rRNA based analysis is the established standard for elucidating microbial community composition. While short read 16S analyses are largely confined to genus-level resolution at best since only a...
2
32
70
Just in time for Thanksgiving, @Covirt19, an international COVID-19 research team that I co-lead along w/@traingene, just launched a COVID-19 Exposure Assessment Tool --> https://t.co/XqFI0ttzvh. Maybe @colbertlateshow & @TheDailyShow will help bring awareness to the public?
0
3
9
Just in time for Thanksgiving, @Covirt19, an international COVID-19 research team that I'm fortunate to co-lead ( https://t.co/KCj0zq49F9) along w/ @AfshinBeheshti , just launched a COVID-19 Exposure Assessment Tool. Download the interactive PDF here --> https://t.co/W3uWEFlvEj.
5
48
100