Todd J Treangen
@traingene
Followers
1K
Following
2K
Media
114
Statuses
1K
Computational tools for metagenomics and public health. Associate Professor at @RiceCompSci After 14 years here time to try something blue: treangen dot 🟦
Houston, TX
Joined September 2009
Absolutely thrilled and honored to receive an @NSF CAREER award from CISE that will support the development of computational approaches for biosecurity & biosurveillance. Special shoutout to the amazing PhD students in my group at @RiceCompSci @Rice_BIOE:
Congrats to @RiceCompSci computational biologist Todd Treangen, who is the fourth Rice Engineering faculty member to win the @NSF CAREER Award this year! The grant will support his development of a computational platform to spot yet-unseen pathogens. 🔗 https://t.co/JWK1KEycJI
8
11
71
Great day in my life!! Finally defended my Ph.D.! I can’t do this without the guidance of my Ph.D. advisor @traingene and my co-advisor @sedlazeck! I’ll soon join @sedlazeck’s group at as an postdoc focusing on cancer genetics and cancer epigenetics.
4
1
17
Rice University's @traingene and @vyao will be hosting the 2nd annual #RADGenomics Workshop at the #AIHealthConference on October 9, 2023. "Reading, Assembling, Analyzing & Designing Genomic Data." View details at https://t.co/AzWVRlPnMx.
1
8
16
Just over a week to go for JXTX Scholarships for @CSHL Genome Informatics! The application process is lightweight and just needs a short statement of goals in addition to your conf abstract. Please help spread the word!
jxtxfoundation.org
JXTX + CSHL 2023 Genome Informatics Scholarships
1
30
19
Our final speaker of #CINEMAParis2023 is Dr. Laurent Moulin from @eaudeparis, talk: “SARS CoV 2 genome monitoring in wastewater by the French OBEPINE network, deciphering the viral dynamics and VOC evolution” Great work on wastewater pathogen monitoring in Paris @lauren_stadler
0
4
8
Day 6 of #CinemaParis2023 is just underway! Great group discussion on relevant research questions for microbiome data led by @kristendcurry. This afternoon will focus on analyzing datasets the participants brought with them & prep for final presentations tomorrow @lauren_stadler
0
0
13
Day 5 of #Cinema2023 was superb! @AlexDilthey and @kristendcurry described techniques for microbiome profiling, then @ZaminIqbal was our invited speaker and gave a great talk on "Using revived 100-year old bacteria and new algorithms to study plasmid evolution" @lauren_stadler
0
0
6
Day 4 of #CinemaParis2023: my former colleague at @institutpasteur, Dr. Sara Vieira-Silva (now Professor at the Johannes Gutenberg University Medical Center in Germany) presenting on gut microbiome, clinical metagenomics and population level studies 🦠🧬@saramavs @lauren_stadler
0
0
10
#CINEMAParis day 3- learning and collaboratively working on environmental microbiome analysis led by @jesethdv ! @traingene @RiceUniversity
0
4
22
Only at #CINEMAParis, live @nanopore sequencing run of a zymo mock microbiome by @Scalene next to lighting talk session🔥 We will dive into bioinformatics methods for environmental and clinical microbiome analysis over the next few days ! (Ps. @Scalene said it was a good run!)
0
3
16
Day 2 of #CINEMA2023 is just getting started at the @RiceUniversity Paris Center!🦉🇫🇷 Today features a full-day, interactive tutorial on microbiome sampling and sequencing 🦠🧬, led by Dr. Joshua Quick (@Scalene), with CINEMA co-director @lauren_stadler moderating todays session
0
2
12
That's a wrap for the first day of CINEMA 2023 ( https://t.co/b8YK4PYGaC) at @RiceUniversity Global Paris Center Dr. Nicola Segata (@nsegata) kicked things off with a wonderful keynote on “Large-scale strain-level computational metagenomics of the human microbiome” On to day 2!
events.rice.edu
A metagenome analysis workshop designed for graduate students, postdoctoral fellows, and investigators from diverse backgrounds and biological inte...
0
3
9
Really excited to see fresh Parsnp at #ASMicrobe , @BKille has been working on some exciting (and long overdue) improvements to Parsnp ( https://t.co/IYSC4mzQj4). More updates soon!
github.com
Parsnp was designed to align the core genome of hundreds to thousands of bacterial genomes within a few minutes to few hours. Input can be both draft assemblies and finished genomes, and output inc...
0
0
9
Great talk by @DrRitaColwell at #ASMMicrobe titled "Cholera in the time of climate change" Dr. Colwell covered over 40 years of research on pathogen🦠 surveillance and ended by highlighting recent study on microbiome analysis for wastewater surveillance: https://t.co/ASW6QMEVxw
journals.asm.org
Traditionally, testing for COVID-19 is done by detecting SARS-CoV-2 in samples collected from nasal swabs and/or saliva. However, SARS-CoV-2 can also be detected in feces of infected individuals.
0
1
6
#ASMicrobe folks if of interest attend my rapid fire talk & poster tomorrow! It focuses on antibiotic resistance detection in wastewater with various approaches! This work is joint first author work with @EstherGeLou2 and great collaboration with the Stadler lab @lauren_stadler
I look forward to attending #ASMicrobe over the next 3 days! Posters/talks from students in my group: antibiotic resistance + nanopore sequencing (@fuyilei96), QuaID (@SapovalNicolae), Olivar (Michael Wang), Parsnp (@BKille), SeqScreen Nano(@AdvaitBalaji), & Squeegee (@beleafsTX)
0
1
5
I look forward to attending #ASMicrobe over the next 3 days! Posters/talks from students in my group: antibiotic resistance + nanopore sequencing (@fuyilei96), QuaID (@SapovalNicolae), Olivar (Michael Wang), Parsnp (@BKille), SeqScreen Nano(@AdvaitBalaji), & Squeegee (@beleafsTX)
0
3
17
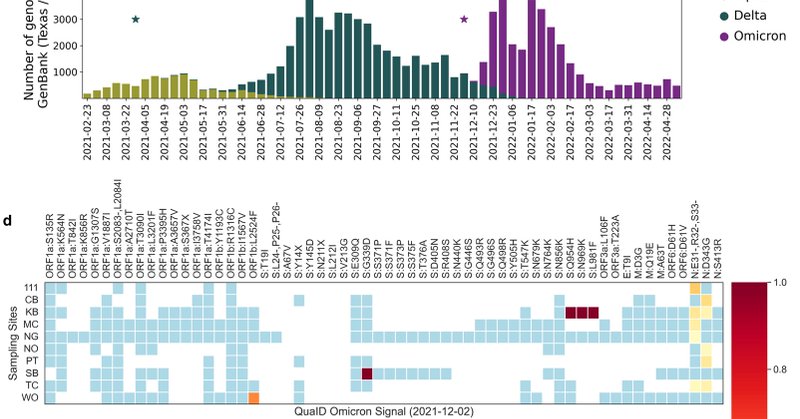
Excited to share that our paper "Enabling accurate and early detection of recently emerged SARS-CoV-2 variants of concern in wastewater", introducing QuaID: a tool for detection of variants of concern in wastewater samples, was published in Nat. Comms https://t.co/SelJKajG3y 1/🧵
nature.com
Nature Communications - Sapoval et al. introduce QuaID, a bioinformatics tool for SARS-CoV-2 variant detection based on quasi-unique mutations. QuaID leverages all mutations, including insertions...
3
26
63
2/2: I'm beyond thrilled to have the opportunity to continue the momentum from the past 5 years and have the opportunity to work alongside truly inspiring students, staff and faculty at an institution with strong leadership & commitment to excellence @RiceEngineering @RiceCompSci
1
0
12
1/2: Overjoyed to share that I was promoted to Associate Professor with #tenure at @RiceUniversity! Profound thanks to my amazing lab members, mentors and collaborators. From a PhD student in Barcelona till now, my journey spans 20 years, 3 countries, 5 languages & 8 institutions
26
10
130
Our latest methods paper "Minmers are a generalization of minimizers that enable unbiased local Jaccard estimation" (aka MashMap3) is out with @BKille @erikgarrison @traingene! There's an interesting backstory here 🧵 \
biorxiv.org
Motivation The Jaccard similarity on k -mer sets has shown to be a convenient proxy for sequence identity. By avoiding expensive base-level alignments and comparing reduced sequence representations,...
5
52
123
4/4 Bonus! If you are attending this year's #nanoporeconf (London Calling 2023), feel free to check out the MethPhaser talk given by the one and only @sedlazeck:
0
2
7