Jon Ipsaro
@jonipsaro
Followers
128
Following
215
Media
10
Statuses
54
Biophysicist who dabbles in swing dancing, sailing, and language learning. Newbie in the twitterverse.
Cold Spring Harbor, NY
Joined December 2016
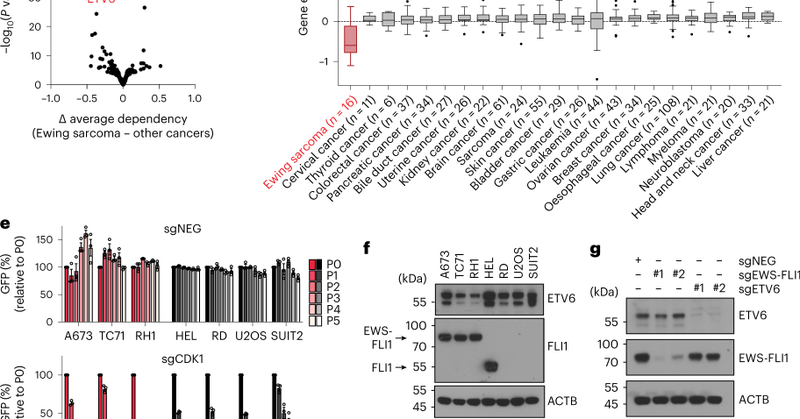
RT @yuangao_yg: Super excited to share our study that uncovers ETV6 as a key growth dependency in Ewing sarcoma! Huge thanks to our amazing….
nature.com
Nature Cell Biology - Independent but complementary studies from Vakoc and Stegmaier identify and characterize a role for ETV6 in counteracting the transcriptional activity of EWS-FLI1 during Ewing...
0
11
0
Sky Wu (吴小丽) @xlw1207 was such a joy to work with on this project. I can't say enough good things about her. Smart, well-rounded, hard working, and fun. A Nature paper and a newly-minted Ph.D! Way to go!.
0
0
18
Had some fantastic interactions presenting at the University of Utah’s RNA interest group! Thanks to @BassLabUtah’s Helen for the smooth planning! Good times!.
0
1
10
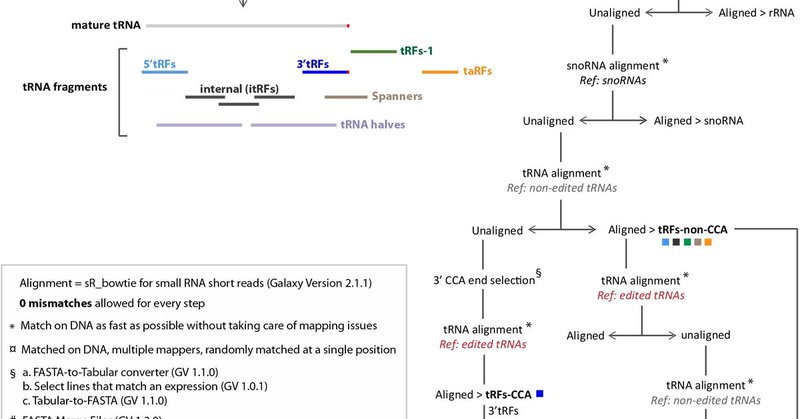
Super excited to give this one a closer look! tRNAs, tRFs, RNA modifications and informatics @anahi_molla 🥳 #100papers #RNA .
frontiersin.org
tRNA fragments (tRFs) are a class of small non-coding RNAs (sncRNAs) derived from tRNAs. tRFs are highly abundant in many cell types including stem cells and...
0
1
3
RT @NobelPrize: BREAKING NEWS: .The 2020 #NobelPrize in Chemistry has been awarded to Emmanuelle Charpentier and Jennifer A. Doudna “for th….
0
23K
0
Just heard about this immunology educational resource in a talk today by @Cheleka_M. Definitely worth a look!.
immunopaedia.org.za
Advancing global immunology education and research. Non-profit, free immunology online courses, clinical case studies, resources and scientific news
0
0
1
Today’s #100papers100days: Bringing RNase activity, RNa binding, and cofactor interactions together in RNAi @BassLabUtah . Heard some of this at #cshlregRNA. Looking forward to reading the whole story!.
0
0
1
Next up for #100papers:.Looking forward to reading this one more closely! @zamore_lab . Terminal Modification, Sequence, and Length Determine Small RNA Stability in Animals
biorxiv.org
In animals, piRNAs, siRNAs, and miRNAs silence transposons, fight viral infections, and regulate gene expression. piRNA biogenesis concludes with 3′ terminal trimming and 2′- O -methylation. Both...
0
0
0
Inspired by @meleckmas, I’m aiming to be more ambitious with my scientific reading this fall. Not sure I’ll be able to match her pace (100 papers in 100 days!) but here’s the first I’m excited to look at from some fellow cryoEM + tRNA + cellular hijacking researchers. 😊🔬.
CryoEM meets RNA! How does a viral RNA’s 3’ end, involved in replication, translation and other processes, use tRNA mimicry and conformational dynamics to coordinate these functions? @Steve_Bonilla‘s preprint of studies of the 55 kDa BMV TLS RNA show how.
0
0
8
At long last on bioRxiv! 😀. This project went from biochemistry to NMR to next-gen seq to custom informatics to cryo-EM. It has been a journey full of challenges and learning. Enjoy! #RNA. Asterix/Gtsf1 links tRNAs and piRNA silencing of retrotransposons
biorxiv.org
The piRNA pathway safeguards genomic integrity by silencing transposable elements in the germline. While Piwi is the central piRNA factor, others including Asterix/Gtsf1 have also been demonstrated...
7
8
52
Also, can we talk about how many of us have an appreciation for good graphic design? The number of informative AND beautiful posters/talks was amazing. #beautifuldata.
0
0
4
Thanks to the organizers, staff, and attendees at #cshlregRNA! Made a lot of new connections and learned so much.👨🏻💻🧬.
1
0
6
The Regulatory & Non-Coding RNA Meeting at CSHL is well underway! Presented this morning and now looking forward to more great sessions. Shout out to all the piRNA folks! #cshlregRNA.
1
1
15
Amazing! If you need a distraction while social distancing, there are some great educational channels here!.
In light of the number of students who are going to be learning from home, Complexly is making its entire catalog of over 2000 educational videos available for free indefinitely. Oh. wait. .
0
1
1