Gilbert Lab
@gilbert_lab
Followers
377
Following
15
Media
0
Statuses
51
The official page of the Gilbert Lab at FSU, where we are interested in the spatiotemporal control of DNA replication and genome architecture
Tallahassee, FL
Joined November 2018
@ScienceMagazine Replication timing maintains the global epigenetic state in human cells. "loss of RIF1 causes near-complete elimination of the RT program ... coupled with widespread alterations in chromatin modifications and genome compartmentalization."
0
18
57
We showed indubitably that DNA replication is required for and upstream of chromatin modification alteration and HiC compartmentalisation disruption. None of this is possible without the help and input from our talented collaborators. Thank you all for your diligent work. 5/5
0
0
2
‘Unaffected' late regions maintain their late timing, which is largely orchestrated by their H3K9me3 association. 'Affected' regions lose their late timing and H3K9me3 binding upon RIF1 KO. Through painstaking cell cycle synchronisation we delineated the order of events. 4/5
1
0
0
Concurrently we found genome-wide reduced H3K27ac and H3K4me3 accompanied by attenuated A compartment interactions by HiC as a result of lost early timing. B compartment was divided into two hubs of interactions according to their response to RIF1KO. 3/5
1
0
0
We disrupted replication timing via RIF1 KO and showed that the nature of timing aberration, rather than being of discrete switches as previously thought, was of dramatically increased heterogeneity in the cell population. 2/5
1
0
1
Congratulations to @KyleKlein7 and @Olivia_Camel on their work published in Science. Many years of hard work went into this amazing story that finally explains why we have replication timing programs - a brief thread. 1/5 https://t.co/PWn9nAGI1G
1
21
82
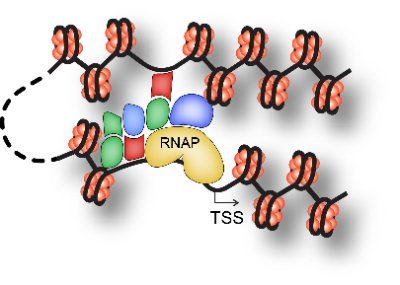
Check out the new biorxiv from graduate student Dan in our lab. He has created a new method (TIP-seq) that combines all of the best aspects of various protein-DNA binding assays to achieve some of the highest resolution data available at the single-cell level.
High throughput genome-wide single cell protein:DNA binding site mapping by targeted insertion of promoters (TIP-seq) [Daniel Bartlett, Vishnu Dileep, Steve Henikoff, David Gilbert] https://t.co/2nnwWoV1Rv ✅~10-fold more unique reads per single cell vs state-of-the-art methods
2
24
137
High throughput genome-wide single cell protein:DNA binding site mapping by targeted insertion of promoters (TIP-seq) [Daniel Bartlett, Vishnu Dileep, Steve Henikoff, David Gilbert] https://t.co/2nnwWoV1Rv ✅~10-fold more unique reads per single cell vs state-of-the-art methods
3
99
321
We cleaned up the Bush 1 mess, we cleaned up the Bush 2 mess and we will clean up this mess. Thanklessly as always!
President Biden will have an unenviable task picking up the pieces & trying to unite the country after this farcical 4-year tragi-comedy of a presidency. The months till inauguration will be dangerous as a desperate Führer in his bunker tries to pull the world down with him.
0
0
1
It’s not - don’t you know - Victor doesn’t believe in TADs!!
0
0
1
High-resolution Repli-Seq defines the temporal choreography of initiation, elongation and termination of replication in mammalian cells
Zhao, Sasaki and @gilbert_lab present a high-resolution study of DNA replication timing in various human and mouse cell lines. Replication patterns generally conserved between cell types, with mid-S phase origins being most variable. https://t.co/99JGQEDbXg
0
5
9
We are also still considering keynote speakers - so if you plan to attend and really have a leader in the field you'd like to hear speak. feel free to offer suggestions. DM me! Architecture people, please RT!
0
1
0
This is early - but everyone studying genome structure and function should keep this meeting on their radar: https://t.co/A9Jbwn9TYq As one of the Co-Chairs, I am happy to answer any questions you may have.
1
2
1