Hanbin Lee
@epigenci
Followers
3K
Following
11K
Media
193
Statuses
5K
Studying how stochastic evolutionary forces shape statistical inference. Statistics PhD student @UMich. 🏳️⚧️
Joined August 2021
New preprint!. We unveil the profound connection between the Ancestral Recombination Graph and Linear Mixed Models. Guess what? Random effects are merely a consequence of mutations running on a fixed genealogy.
biorxiv.org
The ancestral recombination graph (ARG) is a powerful tool for storing and analyzing large genomic datasets, as demonstrated by the ecosystem of software tools taking advantage of the succinct tree...
2
23
65
RT @bookeditor_: 허준이 직접 쓴 동의보감 "초고본"이 발견되었다는 놀라운 소식. 이번 "초고본"은 1613년 찍힌 "초간본"을 출간하기 위해 허준이 직접 집필한 최초의 원고로 보인다고. 와우. 🔍기사 읽기. https://t.c….
khan.co.kr
국보이자 유네스코 세계기록유산인 허준의 <동의보감> 초고본(사진)이 발굴됐다. 기존에는 임진왜란 와중이던 1596년 선조의 명령으로 편찬을 시작해 1613년(광해군 5년)에 처음 찍은 판본인 초간본만 전해졌으며, 허준이 직접 쓴 초고본 존재 자체는 알려지지 않았다. 선조의 지시 이전에 허준이 써놓은 <동의보감>의 틀을 확인할 수 있어 관련 연구에 큰 ...
0
4K
0
RT @AJHGNews: 📣New from Tang & @CharlestonCWKC!.📄A genealogy-based approach for revealing ancestry-specific structures in admixed populatio….
cell.com
We present a method that combines inferred gene genealogical trees with local ancestry calls to estimate ancestry-specific expected genetic relationship matrix (as-eGRM) for admixed populations....
0
8
0
RT @FabrizioRomano: 🚨❤️🤍 BREAKING: Viktor Gyökeres to Arsenal, here we go! Verbal agreement in place between all parties involved. Sportin….
0
54K
0
RT @ErnestRyu: Two cents on AI getting International Math Olympiad (IMO) Gold, from a mathematician. Background:.Last year, Google DeepMin….
0
315
0
RT @NatureRevGenet: New online! An evolutionary continuum between non-coding and coding DNA
nature.com
Nature Reviews Genetics - In this Journal Club, Josué Barrera-Redondo and Susana Coelho recount a 2012 paper by Carvunis et al. that provided a powerful framework for studying de novo gene...
0
17
0
RT @umichkim: Predicting microbial community dynamics is hard: mechanistic models lack flexibility, ML models overfit & ignore physics. Thi….
biorxiv.org
Microbial communities play essential roles in shaping ecosystem functions and predictive modeling frameworks are crucial for understanding, controlling, and harnessing their properties. Competition...
0
4
0
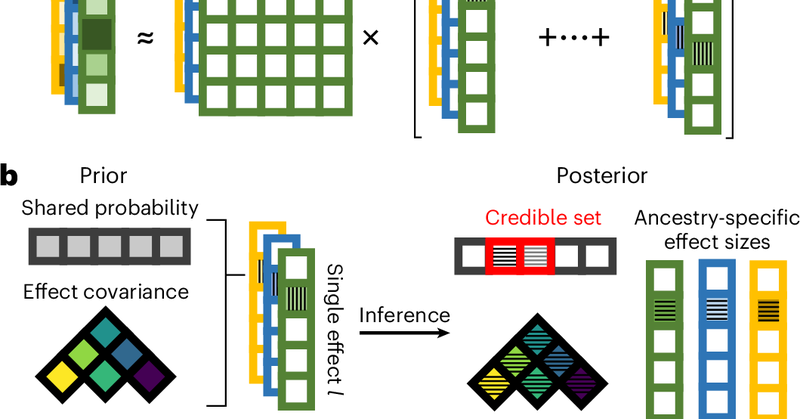
RT @zeyun_lu: Happy to share that our work from the @nmancuso_ lab is out in @NatureGenet! We developed SuShiE, a multiancestry fine-mappin….
nature.com
Nature Genetics - SuShiE is a multiancestry fine-mapping method for molecular quantitative trait loci that leverages linkage disequilibrium heterogeneity to improve resolution, infer cross-ancestry...
0
36
0
RT @ErnestRyu: 7. However, LLMs will become exceedingly powerful for problems that *someone* knows how to solve (in-distribution, in traini….
0
36
0