Christian Mertes
@chr_mertes
Followers
154
Following
668
Media
3
Statuses
240
Scientific researcher @ghga_de and @gagneurlab working on rare diseases, outlier detection, and NGS workflows
Munich, Bavaria
Joined June 2019
Thrilled that our preprint describing our participation in the “SickKids clinical genomes and transcriptomes” @CAGInews challenge is now out in Research Square! One of the winning solutions of the challenge! #Transcriptomics #Genomics.
1
3
16
RT @beryl_bbc: LPSHG is BACK!!. I attended in 2017 & met peers that became lifelong friends & colleagues (@patrick_j_short , @fa7gen). Conn….
0
3
0
RT @gagneurlab: Tomorrow at #ASHG2024, Shubhankar @sg_londhe will present FuncRVP, our model improving rare variant testing by integrating….
0
2
0
RT @Holtkamp_Eva: Feeling extremely honored to receive the #eshg2024 Early Career Poster Award! This work wouldn’t be possible without ever….
0
4
0
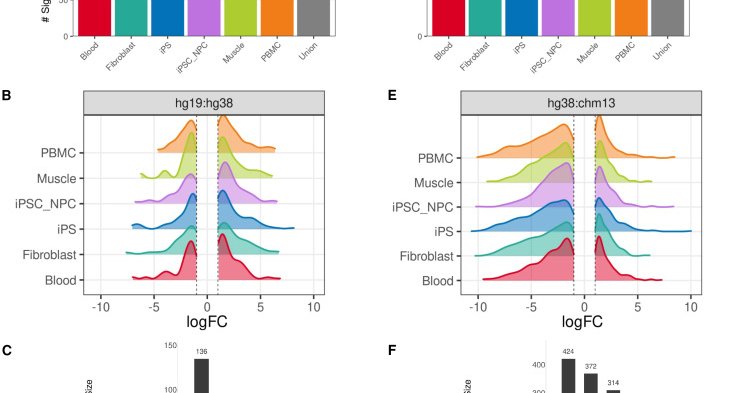
RT @AJHGNews: 📢Online now @AJHGNews!.📰Impact of genome build on RNA-seq interpretation and diagnostics.🧑🤝🧑@raungar @page_goddard @sbmontg….
cell.com
A genome build is the reference sequence to which RNA-sequencing reads are aligned. We found changing the human genome builds (hg19, hg38, and CHM13) impacts interpretation of approximately 39% of...
0
6
0
RT @nickywhiffin: So incredibly happy for super 🌟 DPhil student @quenchentin for winning an early career award for his @eshg2024 presentati….
0
5
0
RT @kubranarci: I am presenting how @GHGA_DE standardize and optimize in-house somatic variant calling workflows using #nextflow. Find me t….
0
3
0
RT @vaym88: Exactly @ahoischen, today at midday at the e poster area or at 1pm at P16.111 we’re showing how RNA-seq has complemented WES, S….
0
4
0
RT @vaym88: Happy to share that DROP v.1.4.0 is out! (our computational pipeline for RNA-seq-based rare disease diagnostics @gagneurlab). I….
0
5
0
RT @gagneurlab: Our study on genomic and transcriptomic aberrations over 3,700 leukemia samples is now published! AbSplice on cancer, driv….
0
7
0
RT @gagneurlab: We have 2 open positions: 1 postdoc and 1 senior scientific software developer /ERC Synergy consortium EPIC, to design and….
0
10
0
Are you passionate about the intersection of human-related #NGS data, biomedical research, and #raredisease diagnostics? The Munich site of @GHGA_DE is expanding with 3 coordinating roles. Please ping me.
0
0
6
RT @CAGInews: The CAGI flagship paper establishes progress and prospects for computational genetic variant interpretation methods. It draws….
genomebiology.biomedcentral.com
Background The Critical Assessment of Genome Interpretation (CAGI) aims to advance the state-of-the-art for computational prediction of genetic variant impact, particularly where relevant to disease....
0
14
0
RT @gagneurlab: When your lecture gets >1,000 students. Exams over multiple TUM campuses for our intro to data science lecture! Thumbs to….
0
3
0
RT @kubranarci: Registration is now open for the @nf_core #nextflow #hackathon from the 18th to 20th of March. Join us for the local node….
nf-co.re
A virtual hackathon with local sites to develop nf-core together
0
6
0
RT @MunichRNA: Hope you are ready for our kick-off meeting #MunichRNA21F! Come and hear the great research by @shu_bhayankar from @ProtzerL….
0
7
0
RT @olcina_lab: Are you a driven postdoc interested in understanding tumour-host interactions to improve paediatric high-grade glioma treat….
0
7
0
RT @raungar: Have you ever wondered how genome build is impacting your RNA-seq results? I am excited to share our new paper on medRxiv inve….
medrxiv.org
Transcriptomics is a powerful tool for unraveling the molecular effects of genetic variants and disease diagnosis. Prior studies have demonstrated that choice of genome build impacts variant interp...
0
33
0
RT @rafalab: 📣A second edition of our Introduction to #DataScience is on its way, now split into two books. After teaching the course this….
0
152
0