Comparative Genomics and Metagenomics Lab
@cgmlaboratory
Followers
449
Following
8
Media
3
Statuses
19
Joined September 2021
Jaime Huerta-Cepas @jhcepas, group leader at @CBGP_Madrid, joint center @La_UPM and @INIA_es, has received the researcher award 2024 of @INIA_es. Congrats Jaime!!
1
6
35
FEBS Fellowships congratulations: we wish Claudia Sanchis López (@clausanchis) from @cgmlaboratory at the Centre for Plant Biotechnology and Genomics, Madrid (@CBGP_Madrid) a rewarding research trip to @EMBLHeidelberg! #FEBSFellowships
https://t.co/hWQ6eS0y0n
2
4
16
"Comparison of gene clustering criteria reveals intrinsic uncertainty in pangenome analyses", in collaboration w/ the Iranzo Lab at @CBGP_Madrid. Nice work derived from Saioa Manzano's master thesis + Yang Liu and Sara González Bodí contribs.@MicroBioBits
https://t.co/mDpE2Jdbnu
genomebiology.biomedcentral.com
Background A key step for comparative genomics is to group open reading frames into functionally and evolutionarily meaningful gene clusters. Gene clustering is complicated by intraspecific duplica...
0
21
52
We are happy to present "TreeProfiler:A command-line tool for computing and visualizing phylogenetic profiles against large trees"( https://t.co/YBtng3UFuo) for annotate and visualize phylogenetic tree with associated metadata... 1/9
1
6
15
Another step in our goal to build a phylogenomics framework that works well with large datasets: TreeProfiler, a handy tool by @deng_ziqi that can map/profile any kind of data against huge trees. Tree + TSV file = auto viz with @etetoolkit v4.0 https://t.co/dDUy5gPzFi
2
51
175
Very exited to discuss the novel gene family analysis we did at @cgmlaboratory in the next edition of the #MVIF !
March #MVIF program is out🤩 Keynote 🇺🇸Prof. Dominguez-Bello (@RutgersU) Talks 🇳🇱@_david_barnett_ 🇩🇪@AnkurMidha3, @vhjarquind 🇪🇸@alvarordr Highlights 🇯🇵Ryza Rynazal 🇮🇹@paolo_manghi 📆Time&date (different from usual) https://t.co/OP4LTcMZpf ✍️Register https://t.co/FxZqztDZOC
1
5
12
our colab paper with the Baker lab is now out!
0
0
14
eggnog 6.0 is out! Lots of great work from the CGMlab crew @anahernandezpl1 Jorge Botas, Carlos P. Cantalapiedra, and @Ginerorama; in close colaboration with the Bork's, von Mering's, and Jensen's labs.
1
20
50
📢 Would you like to work on environmental metagenomics and microbial biodiversity? Our lab has two PhD offers associated to the Spanish FPI program: 1. https://t.co/AoLQc7XnRf 2.
0
8
8
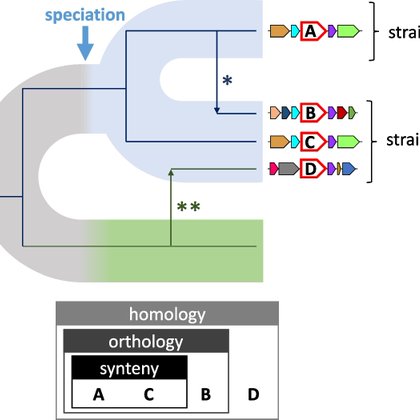
Interested in exploring synteny conservation of prok. eggNOG/KEGG/PFAM groups or genes? Try GeCoViz: genomic context visualisation of prokaryotic genes from a functional and evolutionary perspective super work by @jbotas2 @alvarordr @Ginerorama
https://t.co/UQoJO6zW2v
0
48
170
available 2 postdoc positions for 2 projects starting soon in our lab: 1) BIOUNLOCK: unlocking rare microbial biodiversity in plant soil and phyllosphere. https://t.co/FXXAkAU3q9 2) NUTRIHEAT: root microbiome dynamics on temperature nutrients shock.
0
13
17
We are happy to announce our latest work PhyloCloud( https://t.co/rCiiWzBGQU), an online platform for making sense of phylogenomic data, is now published on @NAR_Open
https://t.co/It3itnkmYh Main features will be unfolded in this thread: @cgmlaboratory @jhcepas @CBGP_Madrid
1
11
22
Our preprint about the discovery and curation of novel microbial protein families from uncultivated prokaryotes is now out. huge work by @alvarordr and many other
Today we release our work on characterizing novel genes from uncultivated prokaryotes! https://t.co/emyTjq1ewc This thread summarizes our main findings: @cgmlaboratory @BigDataBiology @BorkLab @SunagawaLab @CBGP_Madrid @CaixaResearch
0
2
5
glad to share our latest lab paper on the ecological distribution & evolutionary value of 300M genes from 13,000 samples and 14 habitats. https://t.co/L8G6hShF25 w/ @alvarordr @cpcantalapiedra @Ginerorama @BorkLab @luispedrocoelho & more @CBGP_Madrid @INIA_es @CSIC @La_UPM
2
27
56
new collaboration paper from the lab, involving @anahernandezpl1 @cpcantalapiedra. Awesome work led by the Arendt lab at EMBL.
This single-cell analysis of marine sponges led by Detlev Arendt & Jake Musser at @embl provides hints on the possible origin of modern synaptic genes, w/ our modest contrib predicting the evolutionary path of sponge genes. https://t.co/ekEVJLuMAe
@CBGP_Madrid @La_UPM @INIA_es
0
2
5
Next Friday Oct 14 at 12:30 CET I will have the pleasure to host an online CBGP seminar by Manuel Delgado Baquerizo @ManuDelBaq about "The fundamental importance of conserving soil biodiversity". Do not miss it! Link open to everyone (Zoom ID: 867 7585 8243 | 143563)
#SeminarCBGP | 15/10/21 - 12:30h 👨🏽🔬Dr. Manuel Delgado Baquerizo 🌍Universidad Pablo de Olavide, Sevilla. (Spain) 📝The fundamental importance of conserving soil biodiversity 💻Zoom ID: 867 7585 8243 | 143563 #somosUPM
0
10
20
and, another new ms!: @cpcantalapiedra got the new version of eggnog-mapper published: functional annotation, domain mapping and orthology asignments for large genomic and metagenomic datasets. An ongoing collaboration with the @BorkLab @CBGP_madrid
https://t.co/01HDH5tppk
0
27
52
new ms: @clausanchis just got her results published describing ecological vs phylogenetic signal in the distribution of chemoreceptors, reporting a bunch of putative new ligand bind domain families. Great colab work w/ the Lopez-Solanilla lab @CBGP_madrid
https://t.co/Ph4oMVHqHh
0
5
10
The team has spoken and they voted for a lab twitter account, so here go! publication updates, job offers, microbial genomics, phylogenomics, etc. We are: @deng_ziqi @cpcantalapiedra @Calyptrochaeta @anahernandezpl1 @Ginerorama @alvarordr @jbotas2 @clausanchis @jhcepas
2
8
29