Capra Lab
@capra_lab
Followers
591
Following
59
Media
1
Statuses
78
Computers + Evolution + Genomics @UCSF
San Francisco, CA
Joined November 2014
We are hiring! Multiple funded postdoctoral positions are available on projects at the intersection of bioinformatics, evolution, genetics, and electronic health records. Please share!.
3
89
128
I am excited to share our recent work published in @GenomeBiolEvol. We report that human enhancers have both ancient cores and recently evolved DNA. This combo is important for enhancer activity and functional variation across human populations. Check it out!.
How does human enhancer DNA evolve? .Ancient core & younger DNA can combine to form a biochemically active substrate! Distinct TF binding, stronger purifying selection, less common variation hallmark cores v. young DNA. Check out @capra_lab's new work👇 .
0
2
18
RT @CrunchyNeuroSci: Thrilled to share that our beat synchronization GWAS is featured on the September 2022 cover of @NatureHumBehav! This….
0
44
0
RT @CreanzaLab: The genomics of musical beat synchronization out in @NatureHumBehav -- beautiful artwork and a really fun collaboration! @C….
0
4
0
Thanks! Congrats to @Arcane_Asunder!.
In #BMCEcolEvol researchers @capra_lab find examples where the same genetic differences have been maintained in both human and chimp. These are linked to our response to pathogens, body size, alcohol intake, cognitive performance and risk taking behavior.
0
0
2
Check it out and let us know if you have questions. Scores and code available here: . And a web app here:
github.com
COSMIS is a framework for quantifying the mutational constraint on amino acid sites in 3D spatial neighborhoods. The framework currently maps the landscape of 3D mutational constraint on 6.1 amino ...
1
0
2
Led by @computbiolgeek and in collaboration with the @rodendm lab, we used 3D structures from the @rcsbPDB, #AlphaFold, and SwissModel to quantify constraint on 6.1 million amino acid sites in 16,533 human proteins via a COntact Set MISsense tolerance (COSMIS) score.
1
1
1
I am very pleased to (belatedly) share the publication in @NatureComms of our recent work integrating population genetic and protein structural perspectives to quantify mutational constraint in 3D.
nature.com
Nature Communications - Here, Li et al. integrate population genetic and protein structural perspectives to map the landscape of 3D constraint on 80% of human proteins. They show that 3D mutational...
2
37
196
Heroic effort led by @CrunchyNeuroSci to study the genetic architecture of musical rhythm!.
We are very excited to share our newest #musicscience findings on the genetic architecture of musical rhythm, published today in @NatureHumBehav, co-led with @MNiarchou, twitterless Lea Davis, and @NoriJacoby @VUMC_Insights @VUMCgenetics .A thread 1/n.
1
0
5
RT @CrunchyNeuroSci: We are very excited to share our newest #musicscience findings on the genetic architecture of musical rhythm, publishe….
0
38
0
Check out this thread on new work lead by @abin_abe and @Lab_LaBella from the @RokasLab to quantify evolutionary signatures on genomic regions associated with traits from hundreds of GWAS. And @abin_abe has put together a package if you want to try it out yourself!.
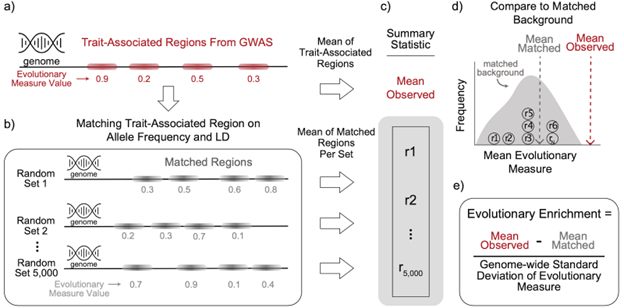
Preprint Alert! .We @abin_abe @Lab_LaBella @RokasLab @capra_lab asked which evolutionary forces have influenced genomic regions associated w/ human traits?.We analyzed >900 GWASs to quantify enrichment for 14 evolutionary measures.Paper: Img: our approach
0
0
7
Check out our lab manuscript template for some tricks, tips, and preferences (in both LaTeX and MS Word). Hoping it can be helpful to others! 🗒️✍️.
Ever sit down to write a manuscript and get feelings of dread? 😰 Hate organizing figure/citation references? 🤬 Try to use #LaTeX but feel immediately overwhelmed? 😵. Check out the @capra_lab manuscript template with tips 📝!!. 1/6
0
5
8
RT @EvonneMcArthur: Experiencing a rollercoaster of emotions since defending my PhD. But these faces capture the best one, pure JOY!!🤗🍾I co….
0
2
0
Check out our preprint and @EvonneMcArthur's thread below. We apply deep learning to reconstruct Neanderthal and Denisovan 3D folding patterns and then ask some cool questions about how this might have affected recent human evolution!.
biorxiv.org
Changes in gene regulation were a major driver of the divergence of archaic hominins (AHs)—Neanderthals and Denisovans—and modern humans (MHs). The three-dimensional (3D) folding of the genome is...
Ever wonder what the #3Dgenome folding🔺of a Neanderthal looked like?! Could it hold any clues as to where and why we have DNA 🧬and trait differences? . Check out our @capra_lab, Pollard lab & @gfudenberg lab collaboration @biorxivpreprint 1/8
0
3
8
RT @benoitbruneau: What? 3D chromatin organization in Neandertal and Denisovan genomes! From my @GladstoneInst and @UCSF colleagues in Tony….
biorxiv.org
Changes in gene regulation were a major driver of the divergence of archaic hominins (AHs)—Neanderthals and Denisovans—and modern humans (MHs). The three-dimensional (3D) folding of the genome is...
0
4
0
RT @colinmbrand: Brilliant new preprint from the @capra_lab and the Pollard lab on the 3D genome organization of a Denisovan and three Nean….
biorxiv.org
Changes in gene regulation were a major driver of the divergence of archaic hominins (AHs)—Neanderthals and Denisovans—and modern humans (MHs). The three-dimensional (3D) folding of the genome is...
0
2
0
RT @cedricboeckx: Nice new work by @EvonneMcArthur @capra_lab and colleagues further probing changes in gene regulation between us and our….
biorxiv.org
Changes in gene regulation were a major driver of the divergence of archaic hominins (AHs)—Neanderthals and Denisovans—and modern humans (MHs). The three-dimensional (3D) folding of the genome is...
0
2
0
Excited to share new preprint on work done by Sarah Fong (@slifong) exploring the evolution of human enhancers with multiple sequence ancestors and relationships to gene regulatory function and selection pressures.
0
2
8
Excited to share our latest work with an amazing team, @ChazinWalter @alexmblee and @ZacNagel lab, on improving tumor #VUS interpretation with an active learning strategy:
biorxiv.org
For precision medicine to reach its full potential for treatment of cancer and other diseases, protein variant effect prediction tools are needed that characterize variants of unknown significance...
0
2
10