Aviezer Lifshitz
@aviezer_l
Followers
56
Following
55
Media
10
Statuses
37
Joined July 2011
RT @NettaMendelson: New paper from the Tanay lab on longevity is out today in @NatureAging! We employ machine learning to tackle a classica….
0
9
0
RT @ymayshar: Our work on rabbit gastrulation by the Stelzer and Tanay labs is now online! Spicing up the classic evo-devo hourglass theory….
0
25
0
RT @ZMukamel: Our work on the functional role of DNMT3A/3B during early development is finally out!.Grateful for the opportunity to work wi….
0
36
0
TET knockouts cause severe gastrulation defects, but in chimeras, they differentiate (almost) completely normally. Check out how @SaiFeng_Cheng and @mittnenzweig use that to separate cis from trans effects!
2
0
4
Congratulations to my sister and brother in-law! @unDtpl9Etkp27U8 @orielber.And to my daughter who is the lead actor!.
The winner of the 2022 Yugo BAFTA Student Film Award for Live Action is “Girl No. 60427,” directed by Shulamit Lifshitz and Oriel Berkovits from The Maaleh Film School in Israel.
0
0
9
Now on Genome Biology: Code: Docs:
github.com
Metacells - Single-cell RNA Sequencing Analysis. Contribute to tanaylab/metacells development by creating an account on GitHub.
We are releasing today a scalable and improved version of #Metacells (MC2), allowing fast and robust analysis of very large-scale single-cell RNA-seq data. See bioarxiv [, code [.
0
0
1
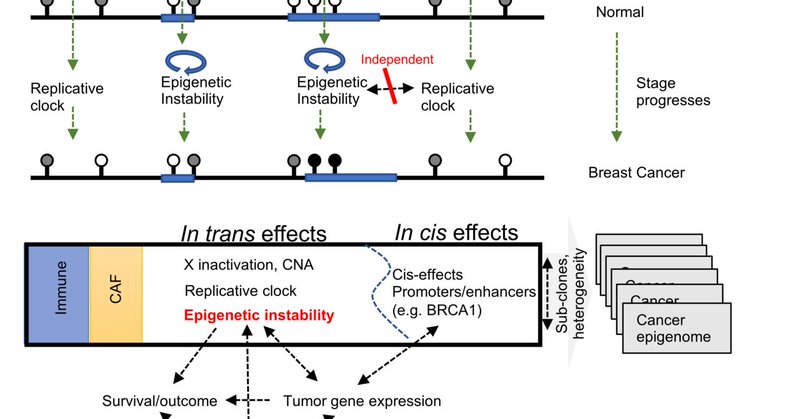
RT @NatureComms: Study investigates DNA methylation landscapes of 1538 breast cancers from the METABRIC cohort and reports epigenomic insta….
0
10
0
Also check out the wonderful blog post by @rnbatra which elaborates more on the emerging model for DNA methylation in cancer: .
communities.springernature.com
What is the impact of DNA methylation alterations in cancer? Is it a cause or consequence of tumorigenesis?
0
2
5
RT @NatureMedicine: In our latest News and Views: Marina Sirota and colleagues @UCSF_BCHSI discuss new data on the application of #machinel….
0
9
0
This layer behaves like a tumor methylation clock. But it is not the common age clock (. And it is not correlated with mutations, gene expression or prognosis. Just a background process slowly degrading the cells’ epigenome.
genomebiology.biomedcentral.com
Background Several recent studies reported aging effects on DNA methylation levels of individual CpG dinucleotides. But it is not yet known whether aging-related consensus modules, in the form of...
1
0
1
𝐅𝐢𝐫𝐬𝐭 𝐥𝐚𝐲𝐞𝐫: most of the genome is losing methylation slowly in a replication dependent fashion. Big blocks that are late replicating lose more methylation (e.g., PMDs . @suscla1 @peterwlaird @benbfly
1
0
1