Adam Broerman
@adam_broerman
Followers
639
Following
594
Media
10
Statuses
31
Dynamic Protein Designer @UWproteindesign
Seattle, Washington
Joined April 2017
Excited to announce our new #proteindesign strategy for allosterically controlling the kinetics of protein-protein interactions! Read on for cool applications in cytokine signaling, biosensing, and protein circuits. https://t.co/jBIHq10Ykd
10
170
770
I'm thrilled to share @adam_broerman's preprint @UWproteindesign about facilitated dissociation in designed proteins. In his designs, a competitor can actively displace a tightly bound partner, speeding up dissociation by several orders of magnitude. https://t.co/7dI9FvHnIS
biorxiv.org
Protein design has focused primarily on the design of ground states, ensuring they are sufficiently low energy to be highly populated[1][1]. Designing the kinetics and dynamics of a system requires,...
1
13
75
This was a massive collaborative effort with @FloPraetorius, @malichtenstein, Christoph and @PiehlerLab, Maxx, Mark, and @StollPatrol, Harry and @DMZuckermanLab, the IPD cores, and all of @UWproteindesign. Huge thanks to all of you!
1
1
11
Also, if you have ideas for how these switchable cytokines can modulate biological systems in new ways, we would love to discuss/collaborate! We envision these tools could open up a whole new way to explore the biology of transient cytokine signaling.
1
0
4
We suspect that the effector folds upon binding, and that this flexibility is key for rapid conformational transitions against resisting force. If you are interested in collaborating to explore the link between flexibility and force generation in these systems, please reach out!
1
0
5
We are really excited to see the new avenues this will open up! If you have ideas that build upon this work and would like to discuss, please reach out. A few we have in mind:
1
0
8
Modulating a previously designed cytokine also demonstrates the generality of our approach: by fusing to our switch, almost any binder can be made to rapidly dissociate from its target!
1
0
8
Christoph @PiehlerLab showed that with this switchable cytokine, adding the effector completely reverses receptor dimerization on the cell surface within 10 seconds and immediately blocks accumulation of phosphoSTAT5.
1
0
10
Finally, the residence time of cytokines stimulating their receptors modulates the downstream response, but native pathways for signal termination are slow, so this is difficult to control. We constructed cytokines which can be disengaged from their receptors far faster.
1
0
10
@malichtenstein and I also used facilitated dissociation to make biosensors which are just as modular as those built previously with our LOCKR platform, but which respond 70 times faster.
1
1
15
This facilitated dissociation behavior can be used to rapidly switch split enzymes and to rapidly release kinetic traps in chain reactions, which would be potentially useful for constructing kinetically governed protein circuits.
1
0
11
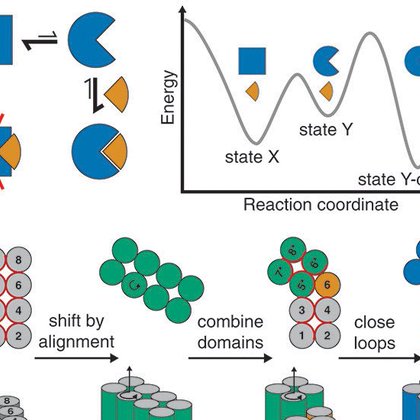
We solved crystal structures of all major equilibrium states of one design, confirming accurate design of these complex multistate proteins. The structure of the strained ternary state(!) shows how the entire complex strains to resolve the designed structural clash.
1
2
35
By designing the magnitude and direction of the induced strain, we could control the partner dissociation kinetics over a wide range. Notably, simple geometric calculations on the AlphaFold predictions of the strained states predicted the dissociation kinetics with an R² of 0.6.
1
0
10
With the partner bound, we measured the base rate of conformational switching and found that the rates of effector binding and subsequent partner dissociation surpass this, confirming an induced-fit mechanism for effector association.
1
0
10
Most efficiently accessing this strained state requires that the conformational switch exerts force in a power stroke. To obtain this behavior, we used a flexible effector and designed an open cleft in the unstrained state where the effector can bind to initiate the switch.
1
0
11
We designed an effector-responsive conformational switch which can be attached to almost any binder to sterically modulate binding its partner. Binding the effector induces a strained intermediate state from which partner dissociation is accelerated by up to thousands-fold.
1
0
14
Protein interaction kinetics often affect the dynamic behavior of biological systems. While high affinity interactions are useful, they usually exchange slowly due to their low off-rates. We developed a general strategy for inducing rapid exchange of high affinity interactions.
1
0
19
If you’re looking for your next position, I can’t recommend this lab enough!
Exciting news part 1: I will be joining @ISTAustria as Assistant Professor and will start my own lab for “Biomolecular Design” in March 2024!
0
1
7
Online at @ScienceMagazine now: my paper with @FloPraetorius at @UWproteindesign We made tiny protein hinges that behave similarly to the hinges you encounter in the macroscopic world (gif credit @ichaydon , protein credit yours truly) https://t.co/hno657zNjV
3
26
162
@definitelyphil and I are excited to see our work on multistate design of hinge proteins finally published as a peer-reviewed paper https://t.co/4T2eHk8g77
science.org
A two-state design of protein switches that couple effector binding to a conformational change is discussed.
1
27
104