ZeitlingerLab
@ZeitlingerLab
Followers
835
Following
200
Media
22
Statuses
134
Our long-term research goal is to understand and predict gene regulation based on DNA sequence information and genome-wide experimental data.
Kansas City, MO
Joined December 2018
Exciting news! Our work on Hippo pathway TF cis-regulation is now published in Cell Genomics! Check it out here: https://t.co/GyI5rFjCoa Grateful to all co-authors who made this possible! @ZeitlingerLab
#HippoPathway #cisregulation #TFcooperativity #Deeplearning
cell.com
Dalal et al. show how AI can be used to systematically uncover the sequence rules that guide how signaling pathways interact with DNA inside cells. This work provides a new lens for discovering the...
2
7
20
We are thrilled to announce the release of Khyati Dalal's (@08Kats05) work, leveraging deep learning and experimental techniques to decipher regulatory mechanisms of Hippo signaling in mTSCs. Congratuations! https://t.co/qYR1QQ1CNE
cell.com
Dalal et al. show how AI can be used to systematically uncover the sequence rules that guide how signaling pathways interact with DNA inside cells. This work provides a new lens for discovering the...
0
6
22
Thanks to our brilliant collaborators @08Kats05 , Charles McAnany, @MelanieWeilert , Cathy McKinney, Sabrina Krueger & thank you to @ZeitlingerLab, @JuliaZeitlinger, and the core facilities at @ScienceStowers. #ResearchTeam #ScienceCollaboration (8/8)
1
0
0
If you want to learn more about the two types of syntax rules we observed (soft and strict syntax) and how they are connected to signaling mechanisms, read our paper. (7/8)
1
0
0
These genome-wide sequence rules and insights allowed us to design small sequence changes that alter the activity of enhancers in vivo, showing that deep learning can learn rules of signaling pathways in a cell-type-specific way. 🧬🔬(6/8)
1
0
2
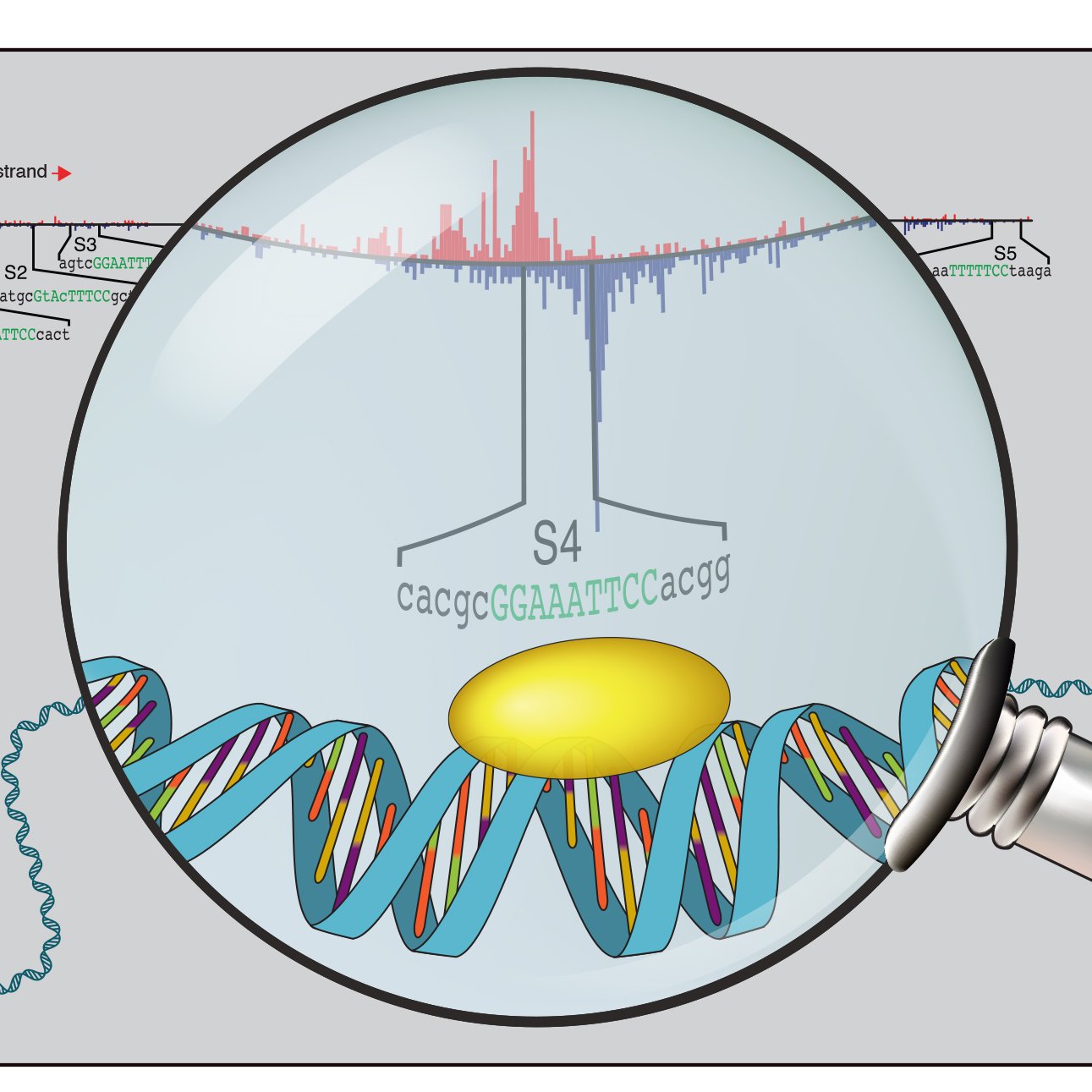
We find that strictly spaced Tead double motifs are widespread, active canonical response elements that mediate cooperativity by promoting labile TEAD4 protein-protein interactions on DNA. Ever heard of protein-protein interactions at the scale of 100 nanoseconds? 🤔 (5/8)
1
0
2
Based on the extracted syntax rules, we validate that the binding of YAP1 can be enhanced, along with TEAD4, by cell-specific transcription factors in a distance-dependent manner, thereby conferring cell-specific response. (4/8)
1
0
2
Next, we found that YAP1 binding is an important determinant for enhancer activation in mouse trophoblast stem cells and driven by the combinatorial rules. (3/8)
1
0
3
Using deep learning on binding data, we extracted sequence rules that drive enhancer activation downstream of the Hippo signaling pathway. We found that binding of the Hippo transcription factors TEAD (DNA-binding) and YAP1 depend on specific motifs of partner TFs (2/8)
1
0
4
In our latest work spearheaded by Khyati Dalal @08Kats05 , we use interpretable deep learning of genomic sequence information to tackle a long-standing question: How do signaling pathways regulate transcription in a cell-type-specific fashion? ( https://t.co/w7jRgpiizi) (1/8)
1
36
124
For a more general summary, check out our press release from @ScienceStowers! (6/6) https://t.co/80kQsneeGr
stowers.org
Stowers scientists are discovering new dynamics for genetic elements during development
0
2
5
Priming of promoters with Paused Pol II by Lola-I reduces the gene activation threshold and increases the transcriptional burst frequency. (5/6)
1
0
1
Lola-I is a promoter pioneer factor that can best access its motif on nucleosomes when located near the edge. (4/6)
1
0
1
Lola-I becomes ubiquitously expressed in late-stage embryos, causing paused polymerases to be loaded onto target promoters in all tissues not just in the tissues where they will be expressed. Thus, priming by Lola-I is surprisingly stage-specific but not tissue-specific. (3/6)
1
0
1
Here we show how a pioneer factor, Lola-I, primes promoters with paused RNA polymerase II during specific stages of embryogenesis. (2/6)
1
0
0
Are promoters just landing pads for the transcriptional machinery? No! They can be key regulatory centers during development. Vivek Ramalingam shows this in some beautiful new work published in @NatureComms. Thread below: (1/6) https://t.co/9cm3e1j47c
1
27
123
We are so proud of @08Kats05 (Khyati Dalal) for passing her defense! Her computational and experimental work is gold-standard for validation of deep learning applications to genomics.
Please join us in congratulating @08Kats05 (Khyati Dalal), @KUMedcenter #graduatestudent in the @JuliaZeitlinger lab, on a successful #thesisdefense! 👏 Well done.
1
2
17
I’m super excited to share our recent work on the establishment of chromatin accessibility during early development! This work is now available online at Developmental Cell. Check it out!
Do only pioneer TFs open chromatin? No! Does motif affinity affect how well pioneering TFs work? Yes! Learn more by checking out our manuscript, now out in Developmental Cell! https://t.co/83YzpoOjZj
4
10
44
Our study on the cis-regulatory code combining deep learning and mechanistic experiments in the early Drosophila embryo is out! Very proud and grateful for everyone’s contribution 😊
Do only pioneer TFs open chromatin? No! Does motif affinity affect how well pioneering TFs work? Yes! Learn more by checking out our manuscript, now out in Developmental Cell! https://t.co/83YzpoOjZj
1
17
114






