Byron Yu
@YuLikeNeuro
Followers
3K
Following
13
Media
32
Statuses
106
Yu Group at Carnegie Mellon University. Neuroscience, brain-computer interfaces, machine learning
Pittsburgh, PA
Joined February 2021
We are excited to share our work on dynamical constraints on neural population activity, published as a cover article in @NatureNeuro. It was led by Emily Oby, @AlanDegenhart, @ErinnGrigsby, with @aaronbatista and team. (1/n).
1
23
91
@MarkChurchland @mjaz_jazlab @FieteGroup @BrunoAverbeck Thank you to @neuroamyo for a wonderful News & Views piece:. (n/n).
www.nature.com
Nature Neuroscience - Our brains evolved to help us rapidly learn new things. But anyone who has put in hours of practice to perfect their tennis serve, only to reach a plateau, can attest that our...
0
0
3
@MarkChurchland @mjaz_jazlab @FieteGroup @BrunoAverbeck Here is the cover image (credit Avesta Rastan):. (8/n)
2
0
3
@MarkChurchland @mjaz_jazlab @FieteGroup @BrunoAverbeck We found that it was difficult for animals to violate their natural activity time courses, thereby providing empirical support for network-level computational mechanisms long-hypothesized by network modeling studies. (7/n)
1
0
1
@MarkChurchland @mjaz_jazlab @FieteGroup @BrunoAverbeck This included asking animals to express the same population activity patterns in the opposite temporal order. (6/n)
1
0
1
@MarkChurchland @mjaz_jazlab @FieteGroup @BrunoAverbeck We used a brain-computer interface (BCI) to encourage animals to override their natural activity time courses. (5/n)
1
0
1
@MarkChurchland @mjaz_jazlab @FieteGroup @BrunoAverbeck We ask to what extent are time courses of the neural population activity (i.e., paths of neural trajectories) “carved out” by constraints imposed by the underlying neural circuity. (4/n)
1
0
1
There has been tantalizing evidence of such principles at play in the brain, including beautiful work from @MarkChurchland, Valerio Mante, @mjaz_jazlab, @FieteGroup, David Anderson, @BrunoAverbeck, and many others. (3/n).
1
0
1
RT @scott_linderman: 📣 The 11th Statistical Analysis of Neuronal Data (SAND11) workshop will be held June 11-13, 2025 in New York at the @F….
0
12
0
We've posted a new paper about how to speed up statistical methods for analyzing multi-area recordings by orders of magnitude:. This work was led by Evren Gokcen, with Anna Jasper (Einstein), Adam Kohn (Einstein), and Christian Machens (Champalimaud).
arxiv.org
Gaussian processes are now commonly used in dimensionality reduction approaches tailored to neuroscience, especially to describe changes in high-dimensional neural activity over time. As recording...
1
6
33
RT @CMUEngineering: Transformative work happens through collaboration. New research from @CMUEngineering & @PittEngineering validates princ….
engineering.cmu.edu
A collaborative team of researchers from Carnegie Mellon University and the University of Pittsburgh designed a clever experiment using a brain-controlled interface to determine whether one-way...
0
4
0
RT @NatureNeuro: Using a brain–computer interface to challenge monkeys to override their natural time courses of neural activity reveals th….
0
20
0
RT @curi_ms: I'm excited to share our #NeurIPS2024 paper with.@jsoldadomagrane @SmithLabNeuro @YuLikeNeuro!.We develop a new brain stimulat….
0
39
0
We encourage you to try SNOPS here:. (n/n).
github.com
Code for SNOPS. Contribute to ShenghaoWu/SpikingNetworkOptimization development by creating an account on GitHub.
0
0
3
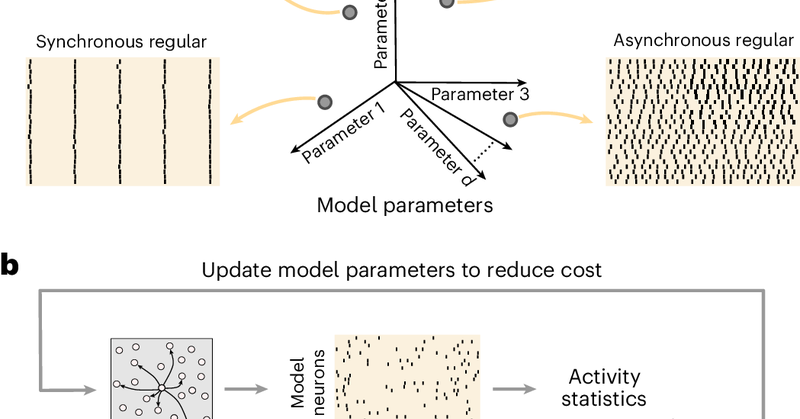
We are excited to share our new method, SNOPS (Spiking Network Optimization using Population Statistics), published in.@NatComputSci. It was led by @Shenghao_W, with @cc_huang11, Adam Snyder, @SmithLabNeuro, and @BrentDoiron. (1/n).
www.nature.com
Nature Computational Science - An automatic framework, SNOPS, is developed for configuring a spiking network model to reproduce neuronal recordings. It is used to discover previously unknown...
3
22
90