Lijia Yu
@YuLijia
Followers
74
Following
35
Media
4
Statuses
40
Joined April 2010
Someone confronted on the spot, and they said “ Maybe there is one, maybe they are common, who knows what. I hope it was an outlier.” Even this explanation is full of implicit racial bias. See the full conv:
dropbox.com
Shared with Dropbox
19
34
709
Remember this habitual offender of racial discrimination. Shame of @MIT 🙃 Hallucination is not a problem of AI, it’s a problem of this group of humans, no? Once we eliminate their corpus from the training set we might solve the hallucination problem:)
7
10
210
I'm shocked to see racism happening in academia again, at the best AI conference @NeurIPSConf. Targeting specific ethnic groups to describe misconduct is inappropriate and unacceptable. @NeurIPSConf must take a stand. We call on Rosalind Picard @MIT @medialab to retract and
133
275
2K
Excited to share our latest publication in single-cell clustering! Check out our paper on "Ensemble deep learning of embeddings for clustering multimodal single-cell omics data" (Now Published!) at
academic.oup.com
AbstractMotivation. Recent advances in multimodal single-cell omics technologies enable multiple modalities of molecular attributes, such as gene expressio
Sorry, I'm late. A new paper of clustering multimodal single cell data is posted at bioRxiv. A huge thanks to @ChunleiLiu0 and my amazing supervisor @PengyiYang82 and @jeanyang21 for the their support and guidance over the course of this project.
0
2
4
Super congrats to Wendy 🎉
0
0
0
Sorry, I'm late. A new paper of clustering multimodal single cell data is posted at bioRxiv. A huge thanks to @ChunleiLiu0 and my amazing supervisor @PengyiYang82 and @jeanyang21 for the their support and guidance over the course of this project.
Continue on her highly successful study on the number of cell type estimation in #singleCell using clustering, @YuLijia led the development of a method for #ensemble deep learning of embeddings for clustering multimodal single-cell omics data https://t.co/tFMxgS3dmN. RT please.
0
0
6
My first preprint is out on bioRxiv!✨ The identification of genes that vary across spatial domains in tissues/cells is an essential step in spatial transcriptomics data analysis. Given this, we evaluate the performance of various proposed SVG methods.
2
4
26
How well do “mini-organs” in a dish recapitulate the human tissue? We set out to answer this question for the human eye, a super fascinating organ! I'm excited to share our preprint on @biorxivpreprint. Check out the full story through the link! 🧵 https://t.co/3501IKJdJB
2
90
426
Excited to share our work @jeanyang21 @PengyiYang82 SimBench @NatureComms, that evaluates #SingleCell simulation methods.
1
12
17
Very excited that our latest work scJoint integrates atlas-scale single-cell RNA-seq and ATAC-seq data with transfer learning is finally out @NatureBiotech! https://t.co/zFK1pkjDHx It is a joint work and a great collaboration between Wong Lab @Stanford and @sydneybioinfo
8
54
185
My final project of my PhD is out in @NatureComputSci!! https://t.co/VDlMj2svdr
#singlecell #cellularidentity #markergenes #differentialstability #Cepo see thread 🧵
6
17
94
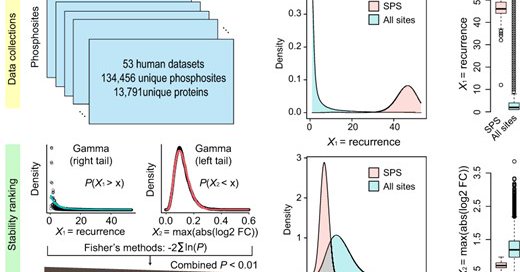
Very excited to share my first first-author paper in @OUPBioinfo !! https://t.co/kv4hPmgkOF Please see thread. #SPS #phosphoproteome #splicesome #cancermutation
academic.oup.com
AbstractMotivation. The advance of mass spectrometry-based technologies enabled the profiling of the phosphoproteomes of a multitude of cell and tissue typ
2
4
10
8/8 We thank our amazing reviewers and editors who helped substantially improve the quality of this work! We will continue benchmarking new methods and new updates on old methods.
0
0
0
7/8 This work would not be possible without my wonderful colleagues at @sydneybioinfo, @SydneyMaths, @CMRI_AUS, @cpc_usyd, @Sydney_Uni. In particular, my partner in SydneyMaths @YueCao16 and our amazing supervisors @PengyiYang82 and @jeanyang21.
1
0
1
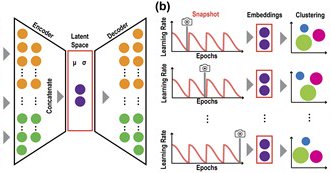
6/8 Source code of scCCESS, our proposed stability-based approach for estimating the number of cell types from scRNA-seq data, can be accessed from
github.com
Single-cell Consensus Clusters of Encoded Subspaces - PYangLab/scCCESS
1
0
0
5/8 We summarised these results into a multi-aspect recommendation to the users and expect this work will foster future development of scRNA-seq data clustering methods.
1
0
0
4/8 Notably, most clustering algorithms tend to under-estimate the number of cell types when the true number of cell types increases in the datasets, whereas interestingly SC3 and Seurat to a lesser degree over-estimate when the true number of cell types increases.
1
0
0
3/8 We found many clustering methods have high instability in estimating the number of cell types. Besides, a higher cell clustering concordance does not necessarily mean a more accurate number of cell type estimation.
1
0
0
2/8 In this study, we set up a systematic benchmark framework for evaluating clustering algorithms on estimating the number of cell types from scRNA-seq data. We design 4 settings by two key aspects: the number of cell types and the cell numbers per cell type.
1
0
0