Wanlu Liu
@WanluLiu
Followers
107
Following
42
Media
0
Statuses
16
Assistant Professor ZJU-UoE institute @zje_institute, Zhejiang University, China. Our lab study epigenetic, genomics & bioinformatics.
Haining, Zhejiang, China
Joined July 2011
We are looking for a postdoc to join a team creating CRISPR-based tools for controlling gene expression by targeting different types of DNA methylation related and non DNA methylation related epigenetic modifications.
6
111
229
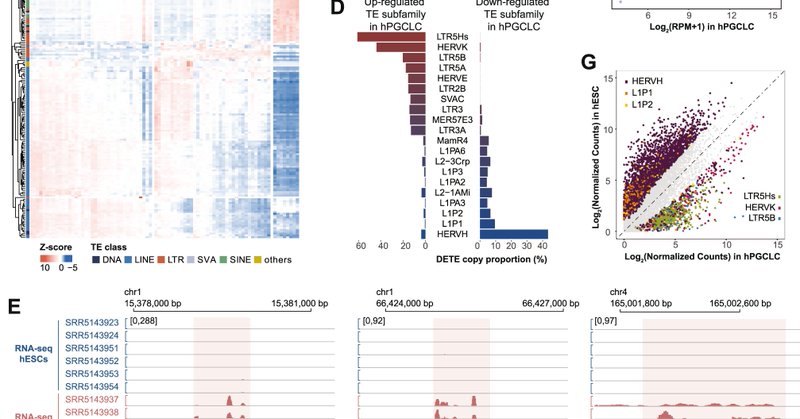
My great honor to collaborate with the Clark lab on this project! Super fun 😀Transposable elements rocks🤟😃
Thrilled to share our latest publication in collaboration with Wanlu Liu’s lab! Human reproduction is regulated by retrotransposons derived from ancient Hominidae-specific viral infections | Nature Communications
0
0
2
Thrilled to share our latest publication in collaboration with Wanlu Liu’s lab! Human reproduction is regulated by retrotransposons derived from ancient Hominidae-specific viral infections | Nature Communications
nature.com
Nature Communications - The transcription factor network required for primordial germ cell (PGC) specification is known to diverge in mammals. Here the authors show that hominidae-specific...
1
60
199
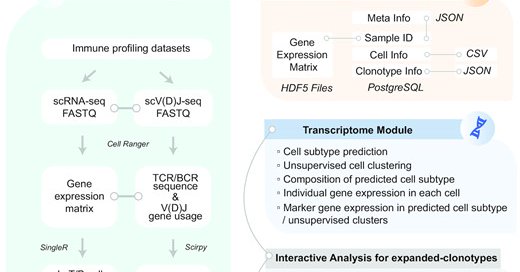
An idea in mind for a long time and now realized by my colleague Wanlu and two brilliant PhD students. huARdb: human Antigen Receptor database for interactive clonotype-transcriptome analysis at the single-cell level
academic.oup.com
Abstract. T-cell receptors (TCRs) and B-cell receptors (BCRs) are critical in recognizing antigens and activating the adaptive immune response. Stochastic
0
1
5
A story started from my PhD, till Postdoc, and finally out after started my own lab for 2 years (p.s. at least it's out before the graduation of my first PhD student). :))) Thanks @JaviGallego80
@SJacobsenUCLA for all the support throughout the journey!!
Finally out! Ectopic targeting of CG DNA methylation in Arabidopsis with the bacterial SssI methyltransferase @WanluLiu @SJacobsenUCLA @IBMCP @UclaMcdb @zje_institute
0
0
8
Our first Bioinformatics paper & my first 'last-author' paper → a nice biologist-friendly web tool(no programming needed) to analyze regions of interest associated DEGs// RAD: a web application to identify region associated differentially expressed genes
0
0
1
[Our new work on PNAS now→check it out]DNA methylation-linked chromatin accessibility affects genomic architecture in Arabidopsis
pnas.org
DNA methylation is a major epigenetic modification found across species and has a profound impact on many biological processes. However, its influe...
0
0
1
RAD: a web application to identify region associated differentially expressed genes https://t.co/DehR8mQB6U
#biorxiv_bioinfo
0
1
1
Check out our new web tool to analyze association of any genomic regions with differential expressed genes! No programming needed, just click the button :) super easy to use 😇 comments and suggestions are welcome
RAD: a web application to identify region associated differentially expressed genes https://t.co/DehR8mQB6U
#biorxiv_bioinfo
0
1
0
I hope you like our new paper on transposon age and non CG DNA methylation https://t.co/I27o5LFBtj feedback will be much appreciated.
4
46
136
After following @biorxivpreprint , finally there is one account that tweets more than @realDonaldTrump and dominants my home page :) Cheers to science 🍺🍺🍺
0
0
0
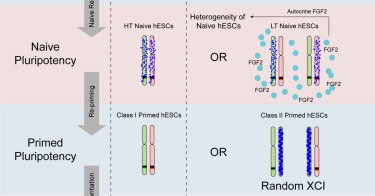
New Resource online now CellStemCell: Overcoming Autocrine FGF Signaling-Induced Heterogeneity in Naive Human ESCs Enables Modeling of Random X Chromosome Inactivation
cell.com
An et al. show that incomplete blockage of endocrine FGF signaling causes heterogeneity of X chromosome status and pluripotent status in naive hESCs. Upon derivation of homogeneous population, naive...
0
2
6
wow
For all colleagues interested in ChIPing hESCs: after sharing in-house with many colleagues @TheCrick we wrote a little methods paper on an optimized ChIP protocol for hESC https://t.co/mz86Km6zj4
0
0
0
Check out our new story collaborating with Dr. William Pastor from McGill University - Naive Human Embryonic Stem Cells Can Give Rise to Cells with a Trophoblast-like Transcriptome and Methylome
0
0
3
NAP1-RELATED PROTEIN1 and 2 negatively regulate H2A.Z abundance in chromatin in Arabidopsis
0
6
8