David Vilchez
@TheVilchezLab
Followers
3K
Following
4K
Media
42
Statuses
1K
Biologist, Professor @CECAD_ @UniCologne @UKKoeln. We study aging, age-related diseases, stem cell biology and proteostasis. @thevilchezlab.bsky.social
CECAD Cologne, Germany
Joined July 2013
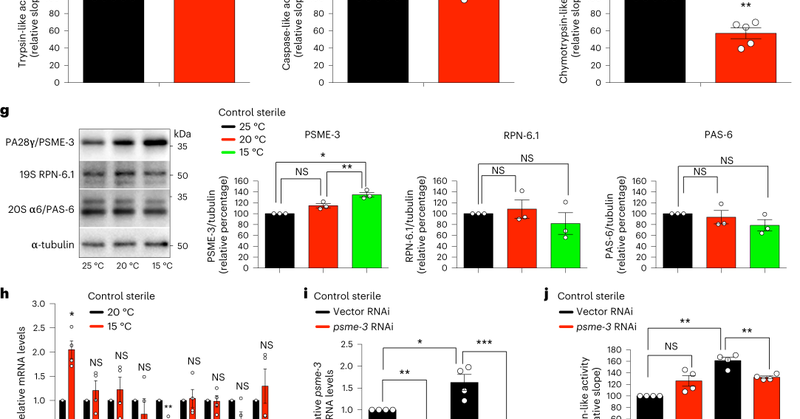
New work from the lab @NatureAging!❄️Lowering body temperature extends #longevity. We find that cold-induced #proteostasis prevents #aging and aggregation of proteins that cause age-related diseases #ALS #HuntingtonDisease in C. elegans and human cells
nature.com
Nature Aging - Moderate cold temperature extends lifespan, but the mechanisms involved are not fully understood. Here, the authors show that moderate cold temperature eliminates aggregation-prone...
28
103
409
A titanic effort from Seda and the lab in collaboration with Nuria Flames’ lab @sedakoyuncu12 @CECAD_ @UniCologne @UKKoeln @IBV_CSIC @NatureAging
0
0
4
We also discovered that the deubiquitinase USP4 leads to EPS8 accumulation during aging. Reducing USP4 prevented EPS8 accumulation, extended lifespan, and lessened disease effects. 👉 Targeting EPS8/USP4 could open new therapeutic strategies for neurodegeneration.
2
0
4
In C. elegans & human cells, we found that age-related EPS8/RAC signaling hyperactivationpromotes pathological aggregation of fifferent proteins (polyQ, FUS, TDP-43). Decreasing EPS8 (or tuning RAC signaling) prevents aggregation & and neurodegeneration.
1
0
1
🚨 New from our lab: Here we explored how aging contributes to harmful protein aggregation in diseases like Huntington’s & ALS. The culprit? An overlooked aging factor — EPS8 and RAC signaling. #Aging #Longevity #Proteostasis #ALSResearch 🧵 https://t.co/Z1kpYZ28GA
nature.com
Nature Aging - Aging is a risk factor for neurodegenerative diseases associated with protein aggregation. Here the authors identify age-related hyperactivation of EPS8/RAC signaling in C. elegans...
4
5
31
New work from the lab! C9orf72 ALS‐causing mutations lead to mislocalization and aggregation of nucleoporin Nup107 into stress granules - Bilican - FEBS Letters - Wiley Online Library
febs.onlinelibrary.wiley.com
Mutations in the C9orf72 gene represent the most common genetic cause of amyotrophic lateral sclerosis (ALS), a fatal neurodegenerative disease. Using patient-derived neurons and C. elegans models,...
1
10
31
We have a 4-year postdoc position open in our Research Unit on proteostasis! The project explores inter-tissue regulation using advanced proteomics approaches. Collaborative & interdisciplinary environment. More details & application here: https://t.co/fe1RfwqwYE
jobs-uk-koeln.onlyfy.jobs
We are looking to support our rapidly growing team as soon as possible:
3
20
37
Excited for the next four years of collaboration on proteostasis research between teams from the University of Cologne, University Hospital of Cologne, and the University of Konstanz
0
0
7
Thrilled to announce that our DFG Research Unit on cell-autonomous regulation of proteostasis has been approved for funding! #aging #proteostasis
https://t.co/BGOf4bhpAu
cecad.uni-koeln.de
A new DFG-funded research group investigates how proteostasis is regulated at the organismal level and its impact on aging and disease.
19
5
73
Happy to share our new preprint, where we find that anle138b reduces Huntingtin inclusions and improves disease phenotypes in #Huntington’s Disease models, showing its potential as an HD treatment. https://t.co/D41LwKgxVo
1
2
17
Don’t miss this opportunity to join Gabriele’s lab! Exciting science!
#Job alert! A #postdoc position in my lab @CECAD_ @UniCologne is open until December 1st. We look for: enthusiasm, curiosity, scientific rigour, and a background in protein quality control, lysosome signalling, or the cell cycle. We offer: fully funded position, top European
0
3
11
Another Nobel Prize for basic research in C. elegans 👏🏼👏🏼👏🏼
This year’s medicine laureates Victor Ambros and Gary Ruvkun studied a relatively unassuming 1 mm long roundworm, C. elegans. Despite its small size, C. elegans possesses many specialised cell types such as nerve and muscle cells also found in larger, more complex animals,
1
4
40
This year’s medicine laureates Victor Ambros and Gary Ruvkun studied a relatively unassuming 1 mm long roundworm, C. elegans. Despite its small size, C. elegans possesses many specialised cell types such as nerve and muscle cells also found in larger, more complex animals,
56
1K
3K
In @elife: Root cap cell corpse clearance limits microbial colonization in Arabidopsis thaliana https://t.co/i6pMs773jg It was a pleasure to contribute to this work from Alga’s lab @Team_Zuccaro and @neto_flames
0
3
13
🔬 Exciting findings from @TheVilchezLab in @CellReports! ALS-mutant FUS variants interact with PARP1 & histone H1.2, driving neurodegeneration. Blocking PARylation or reducing H1.2 may offer new therapeutic hope! 🧠✨ #ALSResearch #Neuroscience 📃➡️
cell.com
Alirzayeva et al. find that ALS-related mutant FUS gains interaction with PARP1 and histone H1.2. Reducing either PARP1 activity or H1.2 levels prevents pathological changes in human motor neurons...
0
5
21
Our study shows that ALS-mutant FUS variants interact with PARP1 and histone H1.2, leading to neurodegeneration. However, inhibiting PARylation or reducing H1.2 levels can prevent disease-related changes in human neurons and C. elegans models #ALS
0
0
6
This was a titanic effort from Hafiza and the lab @CECAD_ @UniCologne @UKKoeln @CGA_age @EKFStiftung @CellReports
1
0
7
New paper from the lab! ALS-FUS mutations cause abnormal PARylation and histone H1.2 interaction, leading to pathological changes: https://t.co/eYWcNqmGXz
#ALS
cell.com
Alirzayeva et al. find that ALS-related mutant FUS gains interaction with PARP1 and histone H1.2. Reducing either PARP1 activity or H1.2 levels prevents pathological changes in human motor neurons...
10
15
73
We have a postdoc/lab manager position available in our lab. The position is initially until December 2025 with the possibility to be extended until December 2032. If interested, please contact me or apply here:
1
98
222