SridharanLab
@SridharanLab
Followers
88
Following
231
Media
11
Statuses
161
Epigenetics, Stem Cells, Development
Joined April 2022
Congrats! Super interesting control of cell identity/ trans differentiation from non-coding variant
Thrilled to share the🥇paper from the lab @atvbahajournals. Here we continue to unravel the mysteries of the 9p21.3 CAD locus showing it drives VSMC transition into an osteochondrogenic state, promoting calcification. @clintomics @UWMadisonCRB
0
0
1
Congrats Erica and Sushmita👏🏽
Big day for our lab with rockstar student @dyneofdata defending her PhD work on matrix factorization variants for regulatory genomics! She is one of a kind combining brilliance and beauty in all her work. #proudadvisor #BDS_PhD
0
0
3
I appreciate Kwangdeok Shin for his dedication in completing this story. I also appreciate other lab members and collaborators, including Dr. Cao, Dr. Wang, Dr. Keles and their groups.
0
1
3
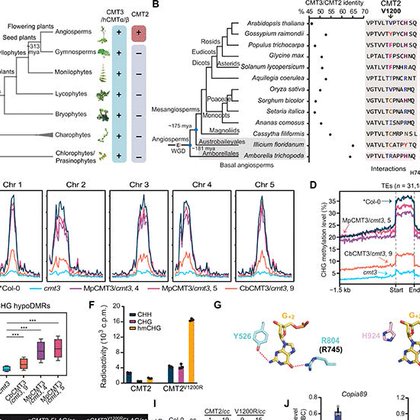
Our work on evolution and functional divergence of DNA methyltransferases is out @ScienceAdvances. Kudos to @jiang_jianjun. We revealed how two closely related CMTs evolve and coordinately shape the non-CG methylation landscapes in plants.
science.org
Functional diversification of substrate-specific DNA methyltransferases accounts for plant genome complexity during evolution.
3
25
78
Delighted to have worked on this perspective on the cis-regulatory code with @JuliaZeitlinger, @JasonErnstLab, @ferhatay, @AMathelier, @AleMedinaRivera, @mahonylab, @sinhasaur
https://t.co/fA8KHcp8Sy. We provide an overview of current progress and some challenges to think about.
arxiv.org
Predicting how genetic variation affects phenotypic outcomes at the organismal, cellular, and molecular levels requires deciphering the cis-regulatory code, the sequence rules by which non-coding...
1
24
83
Congrats Nisha!
We’re kicking off this week as an NIH-funded lab! Beyond excited to receive a HEAL Initiative DP2 New Innovator Award from @NIH_NINDS to create new stem cell tools for region-specific in vitro pain models. Grateful to my incredible lab and @TuftsBME @TuftsEngineer —let’s GO! 🚀
1
0
0
Wisco friends!!! Probably one of the best classes you could take on campus (have seen many of the slidedecks before!) Highly reccommend!
Excited to be co-teaching another edition of Computational Network Biology in Fall2024 with @anthonygitter!
0
1
2
Great to catch up at international society of expt hematology conference with prior trainees -now asst prof at uw-madison, wash u, and assoc prof u Nebraska.
0
3
33
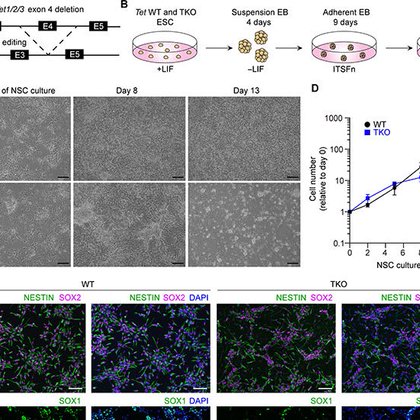
Our work on how Tet-driven DNA demethylation regulates neural specification is published in @ScienceAdvances Hope you find it interesting. Congrats to Ian MacArthur & all authors & @masako_suzuki . @EinsteinMed @Einstein_SCI @EinsteinGenes @EinsteinPhD |
science.org
Developmental DNA demethylation confers identity and gliogenic competence to neural stem cells by commissioning enhancers.
2
10
56
And the best partner to put this meeting together Jamie Lyman Gingerich @UWEauClaire
0
0
0
Heartfelt thanks to our Midwest SDB rep Sharon Amacher , and coorganizers Barak Blum, Matt Gibson, Jeff Hardin, Pascal Lafontant, Anna Sokac, Ahna Skop ( for making a beautiful website) Debora Sinner and Rosa Ventrell
1
0
0
Congratulations to the Wicell presentation award winners Beatriz Ibarra @thisisUIC And Nicole Woodhead @CMB_UWMadison
1
0
1
Congratulations to the Morgridge Institute for Research presentation award winners Emily Adelizzi @uiowa , Harshini Cormaty @uwgenetics And Tracey Porter @NotreDame
1
1
5
Congratulations to the @SocDevBio Award winners Sarah Light @NotreDame, Timothy Nguyen @ UIowaGenetics and Hannah Rice @Mizzou
1
0
0
1
0
2
And the best partner to put this meeting together Jamie Lyman Gingerich @UWEau
1
0
1