Sinan Kilic
@SinKilic
Followers
83
Following
723
Media
2
Statuses
56
Postdoctoral researcher at Center for Protein Research at University og Copenhagen
Joined December 2018
RT @ChoudharyLabCPR: Do enhancers often skip active genes? Our other recent preprint revises current models, that enhancers frequently and….
biorxiv.org
Enhancers play a critical role in regulating transcription. Nearly 90% of human genetic variants identified in genome-wide association studies (GWAS) are located in distal regions, underscoring the...
0
16
0
RT @ChoudharyLabCPR: How do cohesin and CTCF control gene expression and what are their links to enhancers? Our new preprint unravels their….
biorxiv.org
Cohesin and CTCF fold vertebrate genomes into loops and topologically associating domains (TADs). The genome folding by cohesin and CTCF is considered crucial for enhancer-promoter communication and...
0
13
0
RT @ChoudharyLabCPR: Is acetylation of proteins (and histones in particular) a cause or a consequence of gene transcription? We undertook a….
nature.com
Nature Communications - Acetylation of histone lysine residues correlates with transcription activation, but it is currently debated whether histone acetylation is a cause or a consequence of...
0
65
0
RT @Beatfierz: We are hiring! We look for Postdocs in the field of biophysics / chemical biology of gene regulation within a context of a S….
0
31
0
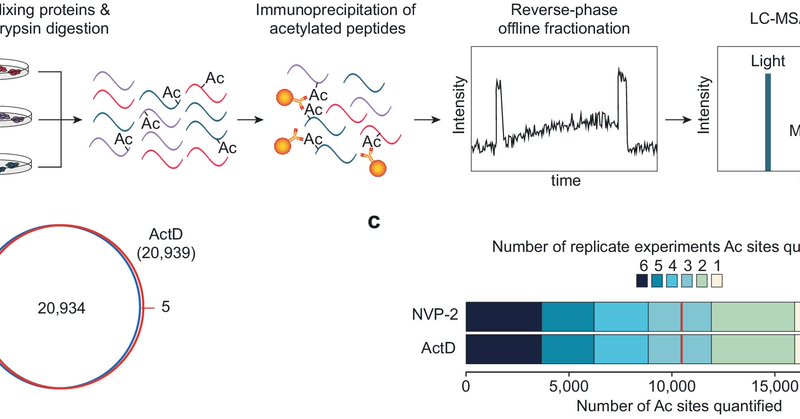
RT @ChoudharyLabCPR: Ubiquitylation occurs broadly in the cell. Have you wondered about the occupancy and dynamics of individual sites on y….
biorxiv.org
Ubiquitylation regulates virtually all proteins and biological processes in a cell. However, the global site-specific occupancy (stoichiometry) and turnover rate of ubiquitylation have never been...
0
52
0
Excited to have been part of this, showing that H2B N-terminal acetylation is a distinct chromatin signature marking active enhancers and CBP/p300-dependent gene promoters Congrats @TakeoNarita80 and Yoshiki!.
H3K27ac has been the go-to hallmark of enhancers for over a decade, but what if another chromatin feature exceeds this in predicting enhancers?.Our paper was just published @NatureGenet Congrats to @TakeoNarita80, Yoshiki and the rest of the team.
0
0
7
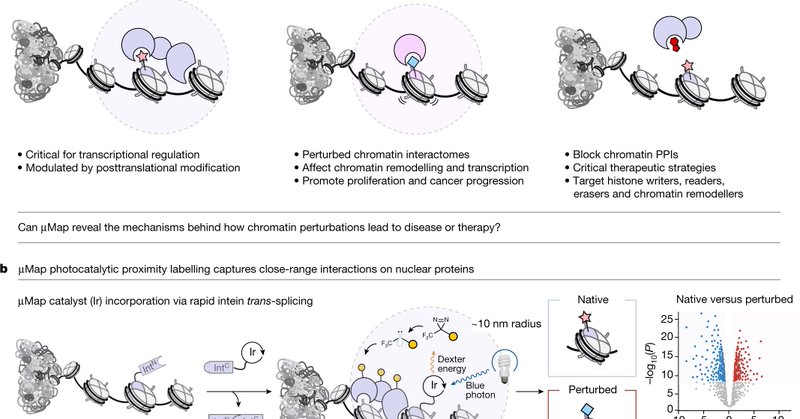
RT @MuirLab: Looking to study chromatin interactomes with proximity labeling? Check out our new paper in @Nature, where we combine our spli….
nature.com
Nature - A proximity labelling-based approach for mapping nuclear protein–protein interactions is described that could advance epigenetic drug discovery.
0
111
0
Impressive cryo-em work, nicely showing how H3-H4 is maintained through transcription, whereas H2A-H2B is semi-conserved. Aligns well with @ChoudharyLabCPR recent pre-print showing the TSS as a bifurcation site for H2A-H2B acetylation retainment upstream, but not downstream.
I am super excited to share our new paper on nucleosome structure dynamics during transcription published in @ScienceMagazine !!.This is a fantastic collaboration with Drs. Ehara and Sekine @riken_en , and @KurumizakaLab . Brief info is in this thread.
0
1
5
Big effort by Takeo and the rest of the @ChoudharyLabCPR for these two preprints on enhancer profiling and cell-type specific gene regulation: .and .
biorxiv.org
Cis-regulatory enhancers are essential for differential expression of developmental and housekeeping genes. However, the specificity of native mammalian enhancers and how it shapes cell-type-specific...
Interested in cell-type specific transcription regulation and enhancer characterization and function? .Check out our two recent preprints and the preview below: .. #Enhancers #Promoters #Gene_regulation #Chromatin @UCPH_CPR
0
0
0
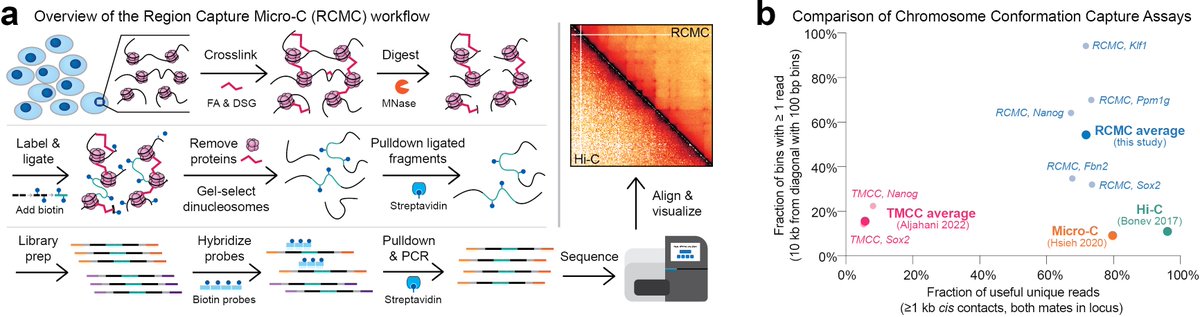
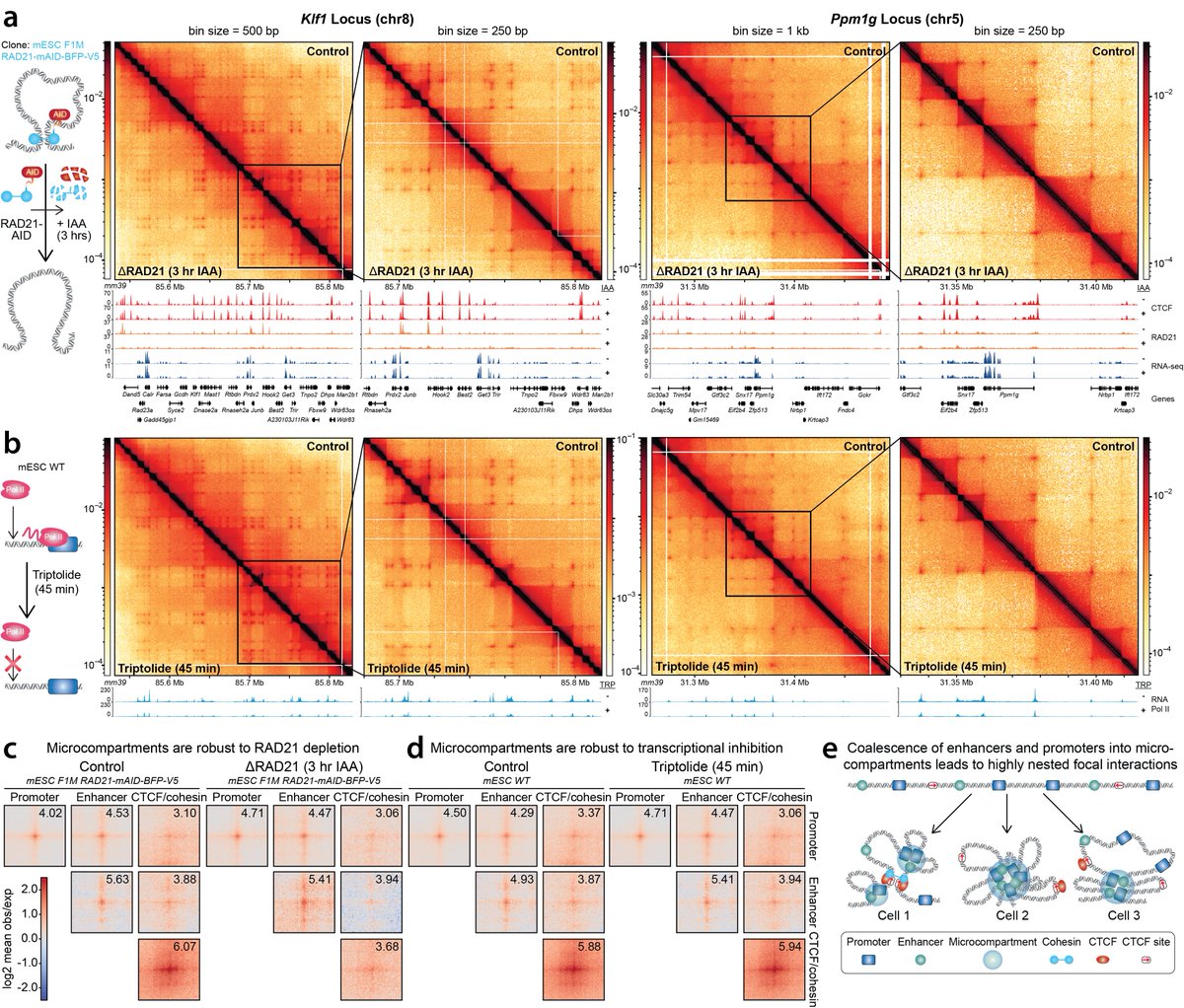
Elegant improvement to Micro-C to provide massively improved depth and identify way more contacts in sub-Mb regions. Interesting that neither transcription nor extrusion contribute much to close contacts. Will be exciting to learn what then does. Congrats to Anders and team!.
(1/n) Excited to share preprint by @ViraatGoel @mileshuseyin. We develop Region-Capture Micro-C (RCMC). By focusing on specific regions, we generate the deepest 3D genome maps thus far. We see coalescence of enhancer and promoters into “microcompartments”.
0
1
1
Excited to see the last paper that I contributed to in a small way during my PhD, out today: Congrats @Heidarsson_lab, Ben and colleagues!.
0
4
39
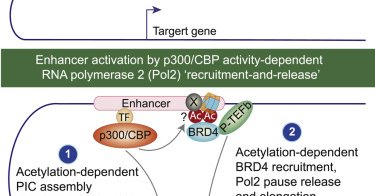
RT @ChoudharyLabCPR: Our paper on the mechanism of p300/CBP activity-dependent kinetic enhancer activation is now out in Mol Cell https://t….
cell.com
A systems-wide analysis reveals that enhancers are activated by p300/CBP-catalyzed acetylation and deactivated by deacetylation. p300/CBP activity promotes pre-initiation complex assembly and RNAPII...
0
1
0
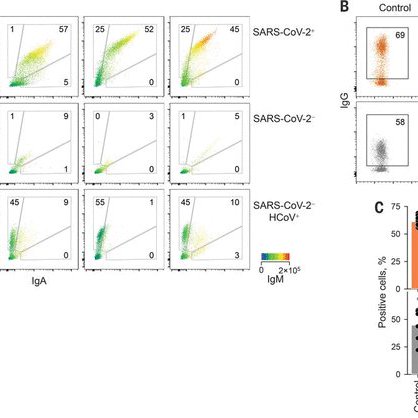
RT @VirusesImmunity: Do some people have cross-reactive antibodies to #SARSCoV2? If so, who are they? And are these cross-reactive Abs prot….
science.org
SARS-CoV-2 neutralizing antibodies can be found in some uninfected individuals—predominantly children and adolescents.
0
380
0
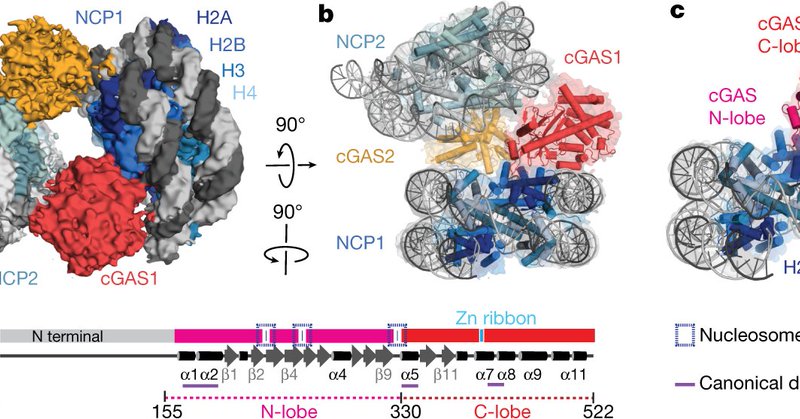
RT @Beatfierz: Check out this new story of @LabThoma, @GaneshRPathare, and the Ablasser Lab @epflSV (and a few nucleosomes from us) on how….
nature.com
Nature - Using cryo-electron microscopy, the authors determine the structure of cGAS bound to nucleosomes and present evidence for the mechanism by which nucleosome binding to cGAS prevents cGAS...
0
8
0
RT @Beatfierz: Great to see this in print @MolecularCell: the chromatin invasion mechanism of the transcription factor Rap1. A great collab….
0
21
0
RT @rbdamgaard: NOW HIRING! I'm looking for fantastic #postdoc and #PhD candidates 👩🔬👨🔬 to join my new lab @DTUtweet in beautiful Copenha….
0
71
0