See-Yeun Ting 陳詩允
@SeeYeunTing
Followers
435
Following
3K
Media
20
Statuses
801
PI in the Institute of Molecular Biology at Academia Sinica. Studying polymicrobial interaction in bacterial community. He/Him
Taipei, Taiwan
Joined April 2011
Excited to learn that our story on characterizing an interbacterial protease toxin is finally out @PLOSBiology "An interbacterial cysteine protease toxin inhibits cell growth by targeting type II DNA topoisomerases GyrB and ParE". Led by my wonderful team @AcadSinica @IMBSinica
3
10
18
🚨 Happy to share our new preprint in which we explore different layers of the phage-bacteria arms race. The highlight: phage homing endonucleases attenuate bacterial defenses 🤯 A 🧵... (1/7) #bacteriology #antiphage #T6SS
@TelAvivUni @TAUMedFaculty
https://t.co/FJUg8Cyr6h
biorxiv.org
The arms race between bacteria and bacteriophages (phages) gave rise to multiple layers of antagonistic mechanisms, many of which remain unexplored. Here, we investigated the anti-phage defense...
1
8
25
Be our IMB colleague! Join us for cool science! OPEN FACULTY POSITION, INSTITUTE OF MOLECULAR BIOLOGY, ACADEMIA SINICA, TAIWAN
nature.com
One tenure-track faculty position is open for a qualified individual to establish an active research program at the IMB.
0
3
5
Check out the new single-tip IMAC-HILIC method from @IPMBSinica for simultaneous enrichment and sequential elution of phosphopeptides and N-glycopeptides in plant samples. Now published in @JProteome_Res! https://t.co/Ib6X2o07wa
0
4
10
Major update to BacFighT6: https://t.co/0UnqXBR6Zf Simulate the role of motility/chemotaxis, capsule production, activation by autoinducers, contact dependent killing, kin-exclusion, foraging and more. Long simulations, extensive data import/export and analysis. Interactive Q&A!
1
6
14
@AcadSinica #ASConference2025 is coming June 30–July 3! 🗓️ Full agenda: https://t.co/CWrCUC9krF
@IPMBSinica is co-organizing 3 interdisciplinary symposia, showcasing our work in plant and microbial biology. Join us for the exciting program!
1
3
5
Programmable cell aggregation by a synthetic biosilicification approach "synthetic multicellular systems via programmable cell aggregation within 15 min. Bio-bricks were created by mixing engineered cells displaying the R5 peptide with sand." https://t.co/FzBXyWjLdZ
3
33
125
Bacteria hitch a ride on neighboring yeast puddles to zoom around. The thin fluid halo surrounding the yeast enables the bacteria to spread more rapidly, turning what appears to be a physical barrier into a stepping stone. More in @BiophysJ: https://t.co/o4UdcQXvrN
1
5
18
Overall, our findings expand our understanding of how bacteria outcompete one another in crowded environments. Check out our story!💪
1
0
0
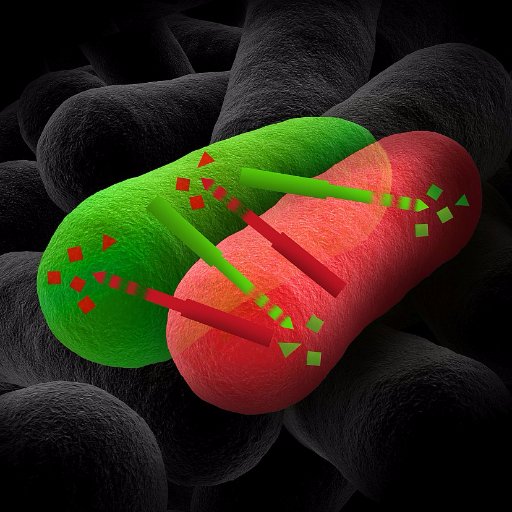
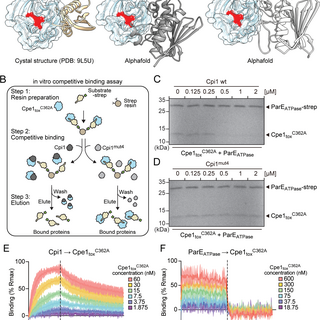
Interestingly, we found that the bacteria secreting Cpe1 are protected from self-poisoning by a protein called Cpi1. Cpi1 blocks the toxin using a unique mode. Unlike many other immunity that block the toxin’s active site, Cpi1 inhibits Cpe1 by blocking the substrate-binding site
1
0
1
Using structural and mass spec approaches, we pinpointed the exact cleavage target sequences of Cpe1: in the short “double-glycine” motifs, specifically LHAGGKF, in GyrB and ParE!
1
0
0
This interbacterial toxin is called Cpe1. It acts like molecular scissors✂️, cutting the ATPase domains of essential topoisomerases, GyrB and ParE, in competing bacteria. Without the critical proteins, the rival cells can't properly copy their genomes, leading to growth stalls.
1
0
0
Link to the story: https://t.co/6fsisGQHgr Bacteria are constantly fighting for survival—not just against bacteriophages, but also against each other. In our new study, we uncover an antibacterial protease toxin that bacteria frequently use to disable their rivals.
journals.plos.org
Bacteria use toxic effectors to outcompete other bacteria, influencing the composition of microbial communities. This study shows that the cysteine protease Cpe1, a widespread toxin in Gram negative...
2
0
1
1/ New preprint! We discovered that Bacteroidales in the gut microbiome can metabolize exogenous DNA 🧬 —turning genetic material into usable nutrients. “DNA-utilization loci enable exogenous DNA metabolism in gut Bacteroidales” 👉
2
9
30
Our new story on #T6SS WHIX effectors and bacterial warfare is out in @PLOSBiology . It has a lot more data than the original bioRxiv preprint, so take a look even if you’ve read that. What did we find? A thread... 1/9 https://t.co/1S69v448Zj
journals.plos.org
The secretion mechanisms of many type VI secretion system (T6SS) effectors in Gram-negative bacteria remain unclear. This study identifies a new class of T6SS effectors, which can harbor either one...
3
8
34
New review out today in Nature Biotech on nanopore-based protein sequencing. Feels... like... something... coming... soon... 👀
6
139
824
We are delighted to welcome Joseph Mougous to the Microbial Sciences Institute. An international leader in microbiology and HHMI Investigator, Mougous joins the faculty @YaleMicroPath later this year.
medicine.yale.edu
Joseph Mougous, an international leader in microbiology and Howard Hughes Medical Institute Investigator, is bringing his lab to Yale School of Medicine.
2
6
54
In summary, our findings show that while phage resistance helps bacteria avoid viral infection, it can come at the cost of losing the battle against other bacteria in the environment. A double-edged sword! ⚔️🔬
0
0
1
More interestingly, even when Salmonella was treated with an enzyme from phages that degrades LPS, it also became vulnerable to bacterial attacks, demonstrating that phages can indirectly weaken bacteria and affect bacteria-bacteria interaction.
1
0
1
Further experiments revealed that the O-antigen, a part of the LPS structure, is crucial for protecting Salmonella against these bacterial attacks. Without it, they became easy targets.
1
0
0