Christopher T Boughter
@Sci_Boughter
Followers
157
Following
544
Media
37
Statuses
225
Ph.D. Scientist studying Computational Biophysics and Molecular Immunology
Cambridge, MA
Joined October 2018
After a good bit of fighting with the code, I'm super excited to finally announce the release of my first ever app! AIMS - An Automated Immune Molecule Separator:
github.com
AIMS - Automated Immune Molecule Separator: An analysis pipeline for distinguishing distinct subsets of Ig and MHC molecules. See below site for documentation - ctboughter/AIMS
1
9
25
A great thread with some much needed testing of ML-based binding predictions! When AF3 can't fall back on its "secret sauce" of leveraging MSA for predictions, it shows just how far we are. Similar to the fundamental issues in TCR/Ab binding predictions. Excited for this paper!.
Here is the Alphafold3 prediction. It is worse than random. High false positive and high false negative (iPTM/PTM shown - other metrics look very similar). 4/n
0
0
5
A great talk by Andreas, strongly recommend it to everyone interested in TCR specificity! Big takeaway for me is that we are quite a ways away from ML being useful in the specificity prediction problem given a random TCR. 99.99% of Andreas' new sequenced TCRs were orphans!.
What are the rules of the immune receptor code? In my talk @KITP_UCSB last Friday I summarised our recent findings, provide a perspective on why ML approaches have so far not achieved breakthrough success, and propose a path forward:.
1
0
6
I was invited to write a textbook chapter for "Methods in Molecular Biology: Immunoproteomics"! The final version will be behind a paywall, so here's a free copy. In it, I outline a step-by-step protocol for using AIMS to analyze immunopeptidomics data.
biorxiv.org
Immunopeptidomics is a growing subfield of proteomics that has the potential to shed new light on a long-neglected aspect of adaptive immunology: a comprehensive understanding of the peptides...
1
2
9
Recently been talking with a lot of people about the shortcomings of AI in biology, especially in T cell biology. Came across a decent summary of some of the issues today: A solid read! I recommend it, even if "crisis" is a bit strong.
nature.com
Nature - Scientists worry that ill-informed use of artificial intelligence is driving a deluge of unreliable or useless research.
1
1
4
Really proud that this paper is finally out today in @eLife! I think a lot of the analysis in this paper can/should change the way we think about a really fundamental aspect of immunology: the interaction between TCRs and MHC.
elifesciences.org
A comprehensive computational study of germline-encoded T cell receptor CDR loops and MHC alpha-helices finds limited evidence for evolutionary conserved interactions, and instead suggests broad...
1
2
13
My first publication as corresponding author is out today in its final form! I'm hoping this methods paper can essentially be the AIMS "bible". If you want to see how we can analyze TCRs, antibodies, peptides, and MHC all in one go, take a look:.
journals.plos.org
Author summary Over the past decade, the success of immunotherapeutics coupled with the declining costs of sequencing have stimulated a near exponential growth in the identification of novel T cell...
0
6
26
RT @CellReports: Biochemical and biophysical characterization of natural polyreactivity in antibodies
cell.com
Binding promiscuity is an inherent property of immune recognition, manifested in antibodies as polyreactivity. Borowska et al. dissect this phenomenon at the molecular level and show that the binding...
0
3
0
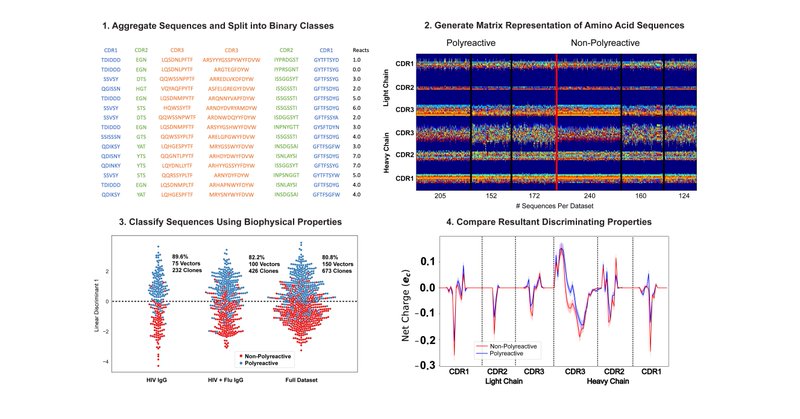
In our bioinformatic analysis of thousands of sequences ( AIMS detected an increase in cross-CDR loop mutual information in polyreactive antibodies. This nicely matches with the increase in inter-loop hydrogen bonds seen in the new crystal structures.
elifesciences.org
A new software developed for high-throughput antibody, T cell receptor, and MHC repertoire analysis uncovers neutrality of the binding interface and intramolecular crosstalk as distinguishing...
1
0
1
My last bit of graduate school-related work is out today in @CellReports ! . Some great experimental work by @mtborowska coupled with some MD simulations really nicely confirmed some of the AIMS predictions we previously published in eLife.
1
0
6
If you're interested in learning more about AIMS, check out the GitHub . Or my recent preprint highlighting the functionality of AIMS. (Which has been in review hell for quite a while, such is life).
github.com
AIMS - Automated Immune Molecule Separator: An analysis pipeline for distinguishing distinct subsets of Ig and MHC molecules. See below site for documentation - ctboughter/AIMS
0
0
0
RT @ImmunityCP: New issue online! Cover depicts work from Esterházy, Brown, @macyrkomnick &co identifying lymph node co-drainage between pa….
0
58
0
RT @andimscience: The QImmuno Lab @iit_ucl @UCL_IPLS is hiring! 🥳 We have an opening for a fully funded PhD position (3 years) on physics-g….
findaphd.com
PhD Project - Physics-guided machine learning of the immune receptor code at University College London, listed on FindAPhD.com
0
26
0