Sara Schulte
@SaraCSchulte
Followers
69
Following
721
Media
7
Statuses
61
MSc computer science @HHU_de | bioinformatics | HLA region | vaccine design | PhD loading ⌛
Joined February 2021
Thrilled about our preprint for @RECOMBconf with @guwekl & @AlexDilthey! We present HOGVAX, a combinatorial optimization approach to peptide vaccine design that exploits overlaps between peptide sequences. https://t.co/nevjPXhe9B
@HHU_de @UniklinikDUS
2
4
5
In conclusion, we show that HLA loss is an important and more frequent mechanism in GBM than previously anticipated. These new insights into how gliomas might escape immune recognition may serve as targets for future personalized therapies.
1
0
0
Through 3D-modeling of individual mutations of TAP1 and B2M, we revealed the potential impact on protein structure.
1
0
0
We found an increased number of mutations in recurrent glioblastoma, suggesting a positive selection or selective expansion of mutagenized tumor cells during tumor progression.
1
0
0
Our analysis identified 46 somatic mutations in the HLA region and associated genes, including eight nonsynonymous mutations. Notably, two of these were in TAP2, one involving an alteration of the start codon.
1
0
0
The pipeline is publicly available on GitHub:
github.com
Snakemake pipeline to call somatic mutations in the HLA region - DiltheyLab/VaCaHLA
1
0
0
Fourth, we designed and applied a custom k-mer filter to eliminate false positive variant calls, followed by manual inspection of the remaining candidates to complete the analysis.
1
0
0
Second, we realign the gene-specific reads separately to canonical references of each gene under investigation. Third, we use three variant callers to perform somatic variant calling.
1
0
0
VaCaHLA takes paired-end Illumina amplicon sequencing data and personalized diploid references as input. First, we align the reads to the personalized reference for a read-to-gene assignment.
1
0
0
We developed a customized #Snakemake pipeline, #VaCaHLA (see first image), to perform somatic variant calling in this highly polymorphic region that is designed to address its inherent challenges.
1
0
0
Here, we systematically investigate the role of somatic mutations in HLA class I and II genes, and genes associated with antigen presentation like B2M, TAP1/2, and MICA/B across isocitrate dehydrogenase wild-type and mutant gliomas.
1
0
0
A previous study indicates an absence of somatic mutations in HLA class I genes in gliomas. However, this study lacked a targeted analysis of HLA class I genes and did not consider class II and further HLA related genes.
1
0
0
Mutations in HLA genes that disrupt antigen presentation are a known mechanism of how tumors can bypass the immune system. Especially malignant gliomas are known for their vicious immune suppression and evasion strategies. However, these strategies are not yet fully uncovered.
1
0
0
The HLA system plays a crucial role in governing the adaptive immune response through antigen presentation. It protects against infectious and non-infectious diseases like cancer.
1
0
0
First things first: Big thanks to @MalteMohme, @AlexDilthey, Gunnar Klau, Wolfgang Peter, Georg Rosenberger, Moritz Schäfer, and so many more people for your tireless efforts throughout the research process to bring this paper to life.
1
0
0
Excited to share our paper on somatic variant calling in HLA genes and genes related to antigen presentation across a cohort of 136 patients with diffuse gliomas published in @CIR_AACR: https://t.co/5NWRoGjXva A thread ... #Bioinformatics #Immunology #CancerResearch
1
3
7
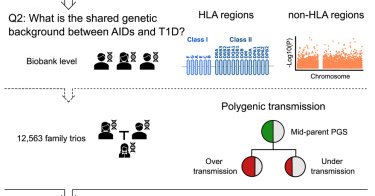
Effects of parental autoimmune diseases on type 1 diabetes in offspring can be partially explained by HLA and non-HLA polymorphisms
cell.com
Leveraging Finnish nationwide multi-generational registers and the biobanks of FinnGen, Wang, Liu, et al. investigated the effect of parental autoimmune diseases on offspring T1D. Their creative and...
0
9
32
How can we ensure that MHC binding prediction algorithms work equitably for patients from all racial and ethnic backgrounds?@PNASNews @Princeton "Toward equitable major histocompatibility complex binding predictions" • Only 180 of 13,000+ human MHC class I alleles have any
0
2
6
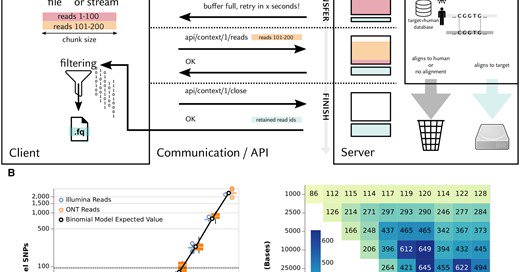
Out now: "SWGTS — a platform for stream-based host DNA depletion" @HHU_de @UniklinikDUS
https://t.co/PTWTdTejp4 Joint work with Philipp Spohr, Max Ried, and Laura Kühle.
academic.oup.com
AbstractMotivation. Microbial sequencing data from clinical samples is often contaminated with human sequences, which have to be removed prior to sharing.
3
5
11
Thrilled :) thanks to the @RECOMBconf committee for the amazing conference and curious folks at my poster for this best poster award
13
3
72