Natalia Pakharukova
@NPakharuk
Followers
33
Following
82
Media
0
Statuses
20
Structural biologist and postdoctoral researcher @Duke & @HHMI. Former @EMBO, @HFSP postdoctoral fellow.
Joined November 2013
Check out our work on the discovery of small molecule modulators of beta-arrestins!
biorxiv.org
β-arrestins (βarrs) are key regulators of G protein-coupled receptors (GPCRs), essential for modulating signaling pathways and physiological processes. While current pharmacological strategies target...
0
1
1
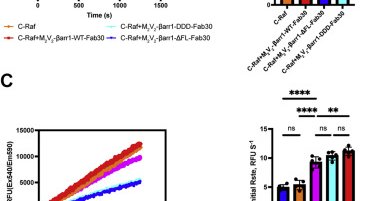
Check out our work on how beta-arrestin activates proto-oncogene kinase Src! This is the first structure of beta-arrestin in complex with the effector enzyme
biorxiv.org
Beta-arrestins (βarrs) are key regulators and transducers of G-protein coupled receptor signaling; however, little is known of how βarrs communicate with their downstream effectors. Here, we use...
0
2
5
RT @biorxiv_biochem: Beta-arrestin 1 mediated Src activation via Src SH3 domain revealed by cryo-electron microscopy .
0
1
0
RT @naturemethods: An iterative approach for improving #AlphaFold protein model quality using experimental density maps, where rebuilt mode….
0
68
0
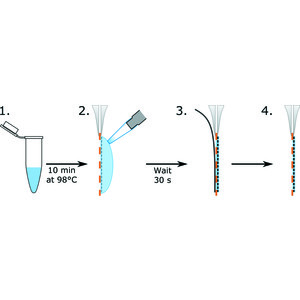
RT @cryoEM_Papers: Polyelectrolyte coating of cryo-EM grids improves lateral distribution and prevents aggregation of macromolecules https:….
journals.iucr.org
Coating cryo-EM grids with single-stranded DNA before applying the sample reduces the aggregation of macromolecules and improves their distribution. It enables the determination of structures from...
0
7
0
RT @cryoEM_Papers: Microsecond melting and revitrification of cryo samples with a correlative light-electron microscopy approach https://t.….
0
8
0
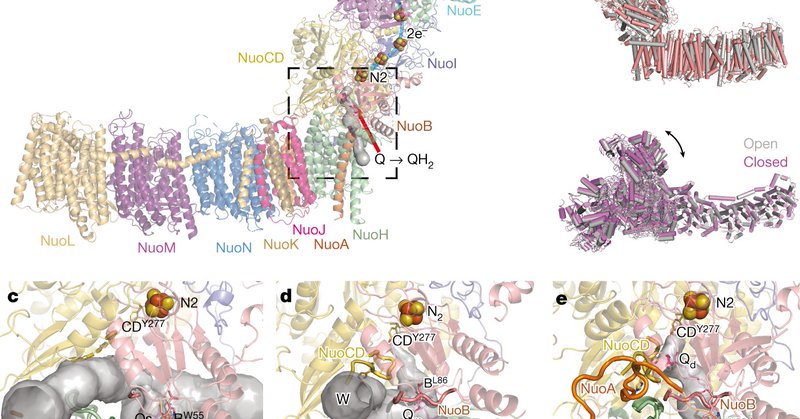
RT @cryoEM_Papers: A universal coupling mechanism of respiratory complex I
nature.com
Nature - Cryo-electron microscopy studies of Escherichia coli complex I suggest a conserved mechanism of coupled proton transfers and electrostatic interactions that result in proton ejection from...
0
7
0
RT @edward_eng: @shen_utah @shen_utah took us through his #cryoEM sample prep journeys with endogenous protein complexes & how https://t.co….
0
2
0
RT @cryoEM_Papers: Automatic and accurate ligand structure determination guided by cryo-electron microscopy maps
0
11
0
Feel proud to be featured by @chngin_the_wrld on our latest paper on #superbugs!.
Today's #shoutout to #womenInSTEM goes to @NPakharuk for her @Nature paper showing how A.baumannii uses archaic chaperone-usher pili to self-secrete into superelastic zigzag springs & attach to #hospital surfaces. #HAI #superbugs #AMR.
0
1
2
RT @cryoEM_Papers: Cryo-EM structure of a type IV secretion system
nature.com
Nature - Cryo-electron microscopy structures of a 2.8 megadalton bacterial type IV secretion system encoded by the plasmid R388 and comprising 92 polypeptides provide insights into the stepwise...
0
16
0
Activated β-arrestin 1 not only scaffolds but also allosterically activates its downstream partners: now shown for C-Raf
jbc.org
G protein–coupled receptors (GPCRs) convert external stimuli into cellular signals through heterotrimeric guanine nucleotide-binding proteins (G-proteins) and β-arrestins (βarrs). In a βarr-dependent...
0
0
0
RT @MedicityL: Read this #preprint on @researchsquare: Archaic chaperone-usher pilus self-secretes into a superelastic zigzag spring archit….
researchsquare.com
Adhesive pili are hair-like appendages assembled via the chaperone-usher pathway (CUP) that mediate host tissue colonization and biofilm formation of Gram-negative bacteria 1-3. Archaic CUP pili, the...
0
2
0
We solved the cryoEM structure of hair-like surface organelles that enable multidrug-resistant bacteria to form biofilms. The study suggested a new approach to treat a wide range of bacterial infections. The paper is under review now and available here:
researchsquare.com
Adhesive pili are hair-like appendages assembled via the chaperone-usher pathway (CUP) that mediate host tissue colonization and biofilm formation of Gram-negative bacteria 1-3. Archaic CUP pili, the...
0
0
0