Michael Desai
@MichaelMDesai
Followers
1K
Following
2
Media
0
Statuses
93
RT @Gautam_Reddy_N: Excited to announce that I've officially joined @PrincetonPhys as an assistant professor. Super grateful to my mentors….
0
14
0
RT @NatureComms: The ability of #SARSCoV2 Omicron to evade immunity is associated with the acquisition of multiple interacting mutations th….
nature.com
Nature Communications - Evolution of the SARS-CoV-2 spike protein is likely driven by many factors, including immune escape and receptor binding. Here, by measuring the binding affinity of more...
0
48
0
RT @NatureComms: The ability of #SARSCoV2 Omicron to evade immunity is associated with the acquisition of multiple interacting mutations th….
nature.com
Nature Communications - Evolution of the SARS-CoV-2 spike protein is likely driven by many factors, including immune escape and receptor binding. Here, by measuring the binding affinity of more...
0
2
0
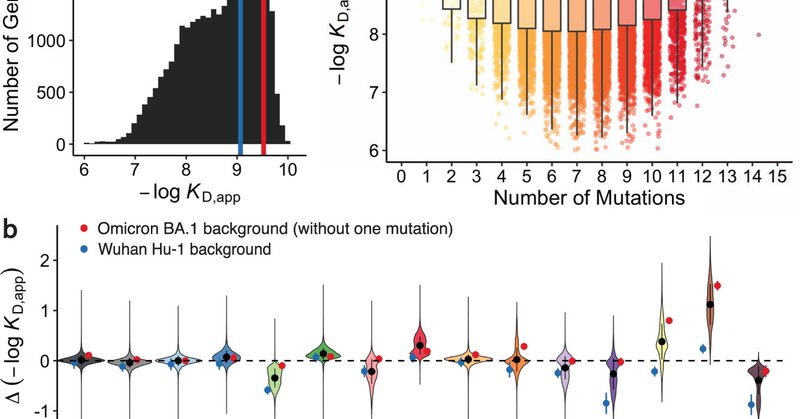
RT @AliefMoulana: Excited to share our work on antibody affinity landscapes of SARS-CoV-2 BA.1--had so much fun working with @ThomasDupic,….
biorxiv.org
The Omicron BA.1 variant of SARS-CoV-2 escapes convalescent sera and monoclonal antibodies that are effective against earlier strains of the virus. This immune evasion is largely a consequence of...
0
2
0
RT @dbweissman: now published: spatial structure can really change the signature of genetic hitchhiking, even in populations that look basi….
academic.oup.com
The reduction of genetic diversity due to hitchhiking is widely used to infer sweeps, but little is known about how space affects hitchhiking. Min et al. m
0
14
0
RT @angphilli: Excited to share our work on epistasis shaping the evolution of SARS-CoV-2 BA.1 — had a blast working on this with @AliefMou….
biorxiv.org
The Omicron BA.1 variant emerged in late 2021 and quickly spread across the world. Compared to the ancestral Wuhan Hu-1 strain and other pre-Omicron SARS-CoV-2 variants, BA.1 has many mutations, a...
0
29
0
RT @_miloj: I recently started poking around with posted datasets using online python notebooks, and have been planning to share them so ot….
0
4
0
RT @angphilli: Thrilled to announce the Phillips lab is opening @UCSF in Sept! We’ll be developing high-throughput methods to examine how m….
0
29
0
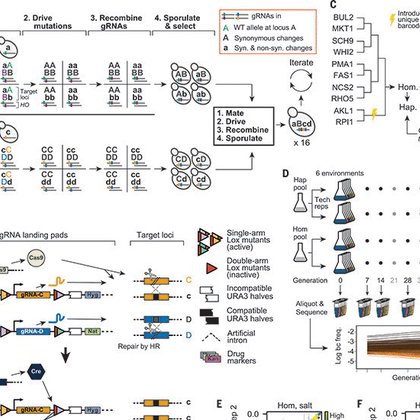
RT @alex_nguyen_ba: Why do some mutations' effects seem to depend on starting fitness? We developed a Cre/Lox CRISPR gene-drive system ==>….
science.org
A genome-spanning fitness landscape reveals how idiosyncratic genetic interactions lead to global epistatic patterns.
0
10
0
RT @AstroKatie: Today in my general relativity class I got to talk about my favorite SUPER WEIRD COSMOLOGY FACT which is that if you have g….
0
1K
0
RT @_miloj: We (@MichaelMDesai and I) just published a preprint about evolution and epistasis! We measured the effects of ~100 insertion mu….
biorxiv.org
As an adapting population traverses the fitness landscape, its local neighborhood (i.e., the collection of fitness effects of single-step mutations) can change shape because of interactions with...
0
23
0
RT @alex_nguyen_ba: Our work, and the monster SI, was a true tour de force by my amazing labmates (Katherine Lawrence and @arturrego_costa)….
0
3
0
RT @alex_nguyen_ba: We analyzed the pleiotropy and epistatic interactions amongst hundreds of QTL per environment from a single cross - hig….
0
1
0
RT @alex_nguyen_ba: Our goal was to approach the scale of GWAS studies that frequently observe these low effect variants, so we used high-t….
0
1
0
RT @alex_nguyen_ba: We developed an experiment to increase the scale of QTL mapping so that we may observe how 'low effect' variants play a….
0
28
0
RT @biorxivpreprint: Barcoded bulk QTL mapping reveals highly polygenic and epistatic architecture of complex traits in yeast https://t.co….
0
3
0
RT @sudiptatung: Excited to share our work on spontaneous whole-genome duplication (autodiploidization) in budding yeast, published in #G3j….
academic.oup.com
Abstract. Spontaneous whole-genome duplication, or autodiploidization, is a common route to adaptation in experimental evolution of haploid budding yeast p
0
11
0