Milo Johnson
@_miloj
Followers
288
Following
431
Media
74
Statuses
390
Postdoc at Berkeley with @bkoskella, @AdamArkinLab, and Adam Deutschbauer, working to measure/predict the effects of mutations in microbes he/him
Joined March 2016
What is the best strategy to win any contest? Eliminate your opponents of course. Recently, my colleague @fernpizza showed how plasmids compete intracellularly (check out his paper just published in Science today!). Together with @baym, we now know how they can fight.
4
29
82
Excited to share pt 2 of my blog on genotype-phenotype maps! We are good at predicting protein sequence->structure. How did it happen? Why was it possible? Can we make similar predictions for protein function, whole-cell phenotypes, or community function https://t.co/CvoFbODtjo
0
0
2
Curious about how evolution takes place? Are you looking for a "first-course" in the subject? My book, Fundamentals of Evolutionary Biology (CRC Press), takes a microbe-first approach and explains how molecular and cellular processes drive evolution.
4
9
37
I'm very excited to share something I've been working on off-and-on for a long time now: a new blog about genotype-phenotype landscapes! The first post is a Gödel-Escher-Bach-style dialogue to introduce the topic. If you like it please share/repost! https://t.co/2l3eswCSzo
1
0
2
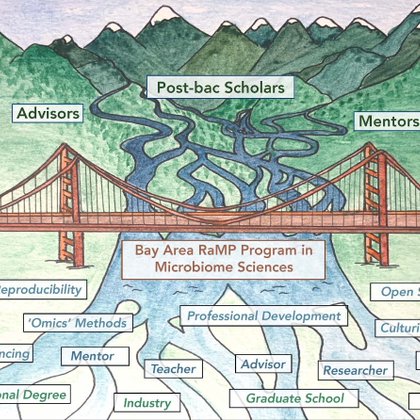
And also also shout out to @bkoskella for making RaMP happen and advising on this project!
0
0
0
Know any recent or upcoming college graduates who are looking for a (full-time&paid!) microbio research experience? Point them towards RaMP! @darian_doakes and I are co-mentors for an MGE project that I think is going to be really cool. Apps+recs by 5/25
sites.google.com
Mission The Bay Area RaMP Program in Microbiome Sciences exists to increase potential for scientific advances by expanding the microbiome research workforce with well-trained, ethical scientists. Our...
1
2
1
Major thanks to Alena for leading the experimental work on this, to Sarah for leading the analysis, and to Sarah and Sergey for carefully pulling it all together, writing, etc.!
1
0
0
All in all, this study gives me more hope that A) there is more low-dimensionality in cellular genotype-phenotype maps than we might think (good news for prediction!), and B) *maybe* someone can figure out mechanistic explanations for these patterns
1
0
0
Sarah and Sergey point out in the supplement that this trend ^ is also predicted by the statistical model proposed by @Gautam_Reddy_N and @MichaelMDesai, which also can broadly be used to understand all of this data.
1
0
1
A last clue: the λe pivot GR parameter is correlated with the average growth rate, so this environment-specific effect is squeezing the adjusted growth rates towards each other (but note the variation away from the line here is real too):
1
0
0
e.g. these knockouts have a fitness cost strictly related to growth rate or ribosome levels, and also have a fixed benefit (reduction in protein production?) that varies based on the environment. Just a hand-wavey example of a guess, I have no idea!
1
0
0
I read this as an indication that there are one or a few (possibly measurable) phenotypes underlying the cellular global epistasis we see in our data, and that at least one is correlated with growth rate, and at least one has some environment-specific basis.
1
0
0
These correlations can apply across environments for many mutations and for the mean of the distribution - only one parameter per environment is needed to shift the background growth rates to a universal scale that can predict effects across environments and mutations.
1
0
0
We can see that it isn't a set property of the genotype - there are environments where background fitnesses are largely uncorrelated, and only the background fitness in the assay environment is good at predicting the mean of the distribution of fitness effects (plot color ~ R^2)
1
0
1
But it's not clear that background fitness / growth rate is the right x-axis - maybe there is something correlated to background fitness that is a more proximal predictor of mutn effects, some unseen phenotype or small set of phenotypes.
1
0
0
We've known for a while that low-dimensional (few-parameter) models can explain some variance in the effects of mutations in proteins or whole cells. This "global epistasis" often emerges in lab microbe experiments as negative correlations bwtn background fitness and mutn effects
1
0
0