Prof. Michael Lin
@MichaelLinLab
Followers
2K
Following
688
Media
245
Statuses
1K
Stanford Neurobiology and Bioengineering Precision molecular design / synbiochem. Also @michaelzlin
Greenberg → Tsien →
Joined July 2022
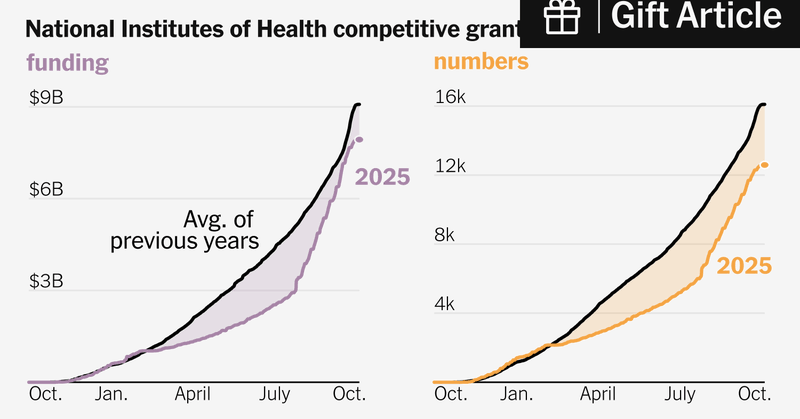
Useful summary of how much has changed this year with NIH/NSF funding. https://t.co/ku8ov7xKNb
nytimes.com
A quiet policy change means the government is making fewer bets on long-term science.
5
33
70
To use an analogy, the belief that voltage imaging is uncommon because the GEVIs haven't been optimized as well as the GECIs is like saying airplane pilots are less common than automobile drivers because less time has been spent engineering airplanes.
1
0
1
@TimothyDuignan The CASP organizers and Nobel committee was very careful not to use the words "protein folding problem". It's important that we don't let people misuse these words in mainstream media. Otherwise, we wont be able to get funding to work on the real problem. 🤪
3
2
33
Eventually this will get cleared up, maybe after we get a chance to present our comparisons between GECIs and GEVIs at different rates. For now though just a reminder that simple explanations are not always 100% correct.
1
0
1
So it's no longer the case that GEVIs are lagging behind in molecular performance because of insufficient engineering. Rather GEVIs have a higher performance bar to clear and will always need better equipment. Still, GEVIs are already better for AP tracking if you can image fast.
1
0
1
The slow imaging rates that GECIs allow are a good fit for the ~20 fps rates of laser-scanning microscopes and CMOS cameras. So GECIs achieving higher usage over GEVIs is due to Ca presenting dual luxuries of abundance and time, allowing big slow signals with common equipment
1
0
0
Specifically, GEVIs need to be much better than GECIs in per-molecule photon change per AP to be usable, because there are >10x fewer GEVI molecules in the membrane than GECIs in the cytosol, and electrical events are >10x faster than Ca, requiring >10x shorter collection.
1
0
0
So why didn't everyone use ASAP3 like they used GCaMP6? Because GEVIs demand high speed and are not as abundant as GECIs, and SNR is related to the square root of both integrated photon count and abundance, so the per-cell SNR is lower for voltage imaging at those faster rates.
1
0
1
But by ASAP3 the GEVIs had already superceded the GCaMPs at the time (GCaMP6f) if you score by photon flux change per molecule per AP. How do we know? Because GCaMP6f gave 20% change per AP, as did ASAP3, but ASAP3 starts at 50% the brightness of GFP while GCaMP6f started at 2%.
1
0
0
GECIs and GEVIs actually started in the same year. Granted, GECIs were easier to improve at first (just add Ca to test). And even GCaMP3, with speed and responsivity only 1/10th of what we have now, was good enough to see some activity in vivo.
1
0
0
Two versions of my famous slide "The Prion Life Cycle": mine vs NanoBanana's version. Obviously, the machines are taking over 😆
3
6
93
And this might seem esoteric now, but after the voltage imaging revolution is complete, it will become common/required knowledge.
1
0
0
Read yet another review today that ascribes GECIs' larger SNR vs GEVIs to their being evolved earlier (suggesting the GECIs are better optimized). This assumption is understandable but incorrect. GEVIs' photonic response per molecule per AP have been as good as GECIs since ASAP3.
1
5
24
Thank you to everyone who came out to #MCCS25 last week! It was great to hear about research happening in the field of molecular and cellular cognition 🧠 🤓 A special thank you to our incredible speakers for sharing their work! Join our mailing list: https://t.co/noAbGJgNHc
0
5
17
Back at UCSD, where I took my first college courses (assembly language and numerical analysis, of all things) and later learned protein engineering in the Tsien lab. The school has grown but is as charming as always. So great to see @jinzhanglab @LoogerL and Nathan Shaner again!
0
2
33
100%. Ridiculous to claim that the biggest breakthroughs in science from here on out will be software. Computing is very important but, beginning with Galileo’s telescope, progress in science has always been driven by technology that allows measurements at new scales.
Francois usually has good takes. But this suggests a bit of cluelessness about what the key barrier to progress in biology is. It's not algorithms or AI. It's still a lack of the ability to measure many important things in cells ie. assay techdev. Perturb-seq is not all u need.
5
8
117