Lucas Arnoldt
@Lucas_Arnoldt
Followers
261
Following
1K
Media
6
Statuses
111
PhD student @ Helmholtz Munich | Interested in Genetics, Omics, Digital Health, Deep Learning.
Munich
Joined March 2016
Current multimodal single-cell integration methods act as black boxes, lacking meaningful interpretability. We introduce NetworkVI, a VAE that performs integration via gene-gene interactions and the Gene Ontology for biologically grounded analysis.
biorxiv.org
Multi-omics technologies allow for a detailed characterization of cell types and states across multiple omics layers, helping to identify features that differentiate biological conditions, such as...
2
6
11
RT @razoralign: NetworkVI: Biologically Guided Variational Inference for Interpretable Multimodal Single-Cell Integration and Mechanistic D….
0
8
0
NetworkVI is a group effort by Julius Upmeier zu Belzen, Luis Herrmann, Khue Nguyen, supervised by @fabian_theis & @nebww & @CaptainSysBio at @ChariteBerlin and @HelmholtzMunich. Code: .Preprint: .🧠 Open-source & ready to use!.
biorxiv.org
Multi-omics technologies allow for a detailed characterization of cell types and states across multiple omics layers, helping to identify features that differentiate biological conditions, such as...
0
3
8
RT @Tillmann_Rhe: Very happy to see this online after the presentation at the @imimicworkshop last year! .In this paper, we propose Seg-Hi….
0
1
0
RT @NobelPrize: BREAKING NEWS.The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to….
0
9K
0
RT @sid_thesci_kid: new paper drop in @NatureBiotech .It was a journey and half getting this out, and I couldn’t have done it w/o the kinde….
0
11
0
This project was co-led by @sid_thesci_kid, @JacobMGershon, and Jeremiah Sims, from whom I learned a lot and am very grateful for the work we did together at @UWproteindesign.
0
0
1
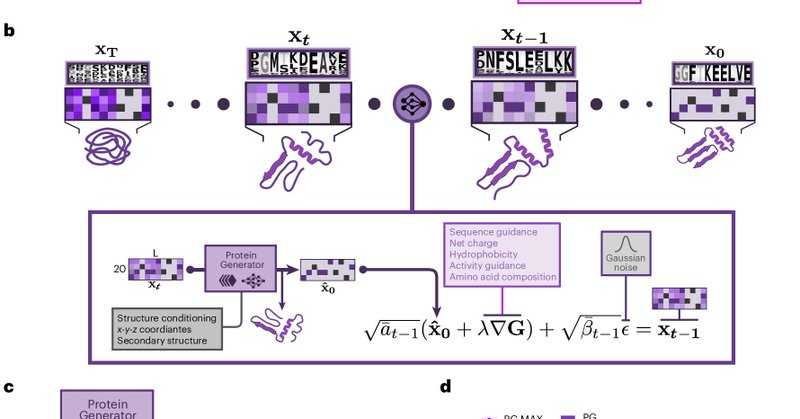
We demonstrate how joint modeling of sequence and structure, can be employed for scaffold design, multistate design, and activity guidance, among other things. Please also check out Sam’s thread, which shows so much more cool stuff.
🥁 Our manuscript "Multistate and functional protein design using RoseTTAFold sequence space diffusion" is finally out @NatureBiotech! 🧵b/c it's changed a LOT since preprint! . New highlights: . - High res structures. - Conditionally caged peptides. - Multistate design
1
0
1
Thrilled to share our @NatureBiotech paper “Multistate and functional protein design using RoseTTAFold sequence space diffusion”
nature.com
Nature Biotechnology - ProteinGenerator simultaneously generates protein sequences and structures using sequence space diffusion.
1
12
50
RT @NatureBiotech: Multistate and functional protein design using RoseTTAFold sequence space diffusion https://t.co….
0
54
0
RT @Jas_Hughes: The fact that there are multiple candidates for what this linear regression is blows my mind 🤯. (Original, of course: https….
0
2K
0