Kai
@KaiPeiLi
Followers
27
Following
169
Media
4
Statuses
24
Serious science for fun.
Ann Arbor, MI
Joined March 2019
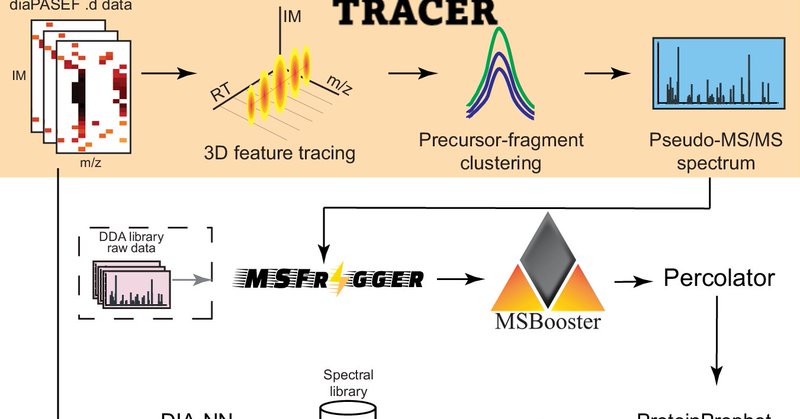
.@KaiPeiLi @nesvilab @fcyucn present diaTracer, a spectrum-centric tool for diaPASEF data, enabling direct peptide identification and quantification without spectral libraries. #BiotechNatureComms
https://t.co/pLTEUyyjwb
nature.com
Nature Communications - Data-independent acquisition advances proteomics quantification. Here, the authors present diaTracer, a spectrum-centric tool for diaPASEF data that supports broad...
0
3
13
Come to my poster at HUPO on Tuesday! Direct spectrum-centric analysis of diaPASEF data using diaTracer and #FragPipe. See how well it works for spatial proteomics, CSF, plasma, HLA, PTMs and more! Updated preprint: https://t.co/pNEXItzC6R. With co-authors Kai, Fengchao, Guo Ci.
2
3
52
Introducing #diaTracer for diaPASEF data! Extending our DIA-Umpire concept to IM data it performs 3D (m/z, IM, RT) processing to generate DDA-like “pseudo-MS/MS” spectra, enabling 'library-free' analyses including PTM, nonspecific, and even open searches! https://t.co/NCGvvSbAfJ
0
11
72
Friends, #FragPipe 22 has been released, and it's a big update! diaTracer enables spectrum-centric analysis of diaPASEF data. Skyline integration. Koina server for more deep-learning prediction options. DDA+ mode for ddaPASEF. DIA glycoproteomics and new chemoproteomics workflows
5
26
136
PLDMS: Phosphopeptide Library Dephosphorylation Followed by Mass Spectrometry Analysis to Determine the Specificity of Phosphatases for Dephosphorylation Site Sequences | https://t.co/n8fLYfR6oV
#proteomics
1
1
1
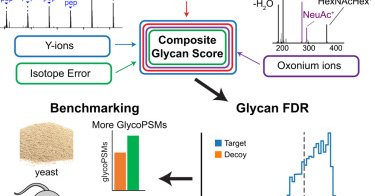
Are you a fan of delicious PTMs? On the day when the Wordle's word was SUGAR came out our manuscript in MCP led by @DanPolasky on FDR control for glycan assignments in glycoproteomics. Fully implemented in #FragPipe as part of MSFragger Glyco workflows!
mcponline.org
In BriefGlycoproteomics has seen rapid advances in methods for identifying glycopeptides, but challenges remain confidently determining the composition and structure of the attached glycan. We have...
1
18
89
Large-scale integration of the plasma proteome with genetics and disease @NatureGenet : “Combining #proteomics, #genomics and #transcriptomics to improve our understanding of disease pathogenesis and assist with drug discovery and development.”
0
6
20
The benchmark metrics demonstrated in this study will enable users to select computational pipelines and parameters for routine analysis of phosphoproteomics data and will offer guidance for developers to improve computational methods ― @bcmhouston @UMich
https://t.co/xhCNwR4sZM
0
3
2
It took "only" 1 year and 11 months to get it published but our work is finally in Nature Communications: https://t.co/3Iq6GL4g2K
@AshrafBrik @IndrajitSahu20 @aleph_reish
nature.com
Nature Communications - The 20S particle is part of the 26S proteasome, but also exists as a free complex. Here, the authors outline signature activities of the 20S and combine chemical,...
4
9
41
Welcome to pGlyco3! pGlyco3 is my favorite project in my ten years of computational proteomics. I am glad to share it with every user. #glycotime
https://t.co/HB6FWJ2KSC Thank all collaborators and all users for using and testing pGlyco3.
biorxiv.org
We presented a glycan-first glycopeptide search engine, pGlyco3, to comprehensively analyze intact N- and O-glycopeptides, including glycopeptides with monosaccharide-modifications. We developed an...
2
11
30
Our second manuscript on ethics in proteomics is available @molcellprot. As proteomics is poised to take an important role in the clinic, together with Gilbert Omenn and @MannPorsdam, we review and discuss ethical principles, opportunities and constraints https://t.co/4Ow5WPrzgS
2
20
66
Genetics meets proteomics: perspectives for large population-based studies | https://t.co/0MY0N0ipEi
#proteomics
1
4
20
DeepRescore was selected as the front cover in Proteomics latest issue ( https://t.co/5egqBZK72n). Thanks to everyone's work on this project. Thanks to my friend Miaoyu Li for designing this cover with me. (Two AI robots work hard to recover useful IDs from discarded results.🤖🤖)
2
0
2
🔴 𝗕𝗔𝗬𝗘𝗥𝗡 𝗔𝗥𝗘 𝗖𝗛𝗔𝗠𝗣𝗜𝗢𝗡𝗦 𝗢𝗙 𝗘𝗨𝗥𝗢𝗣𝗘! 👏👏👏 #UCLfinal
506
7K
35K
UNBELIEVABLE! UNBELIEVABLE! UNBELIEVABLE! GO! BAYERN! GO!
0
0
0
Really love this webinar! And thank you a lot for supporting PDV all the time.
Thanks to everyone who attended my webinar on MSFragger today. In support of all international scientists and students, I put a little flag next to the picture of each person in my lab who contrubuted. There were 7 flags, and my lab is not that big. Science should have no borders
0
0
1
The gnomAD package is officially out! We look at those genes where we see loss-of-function variants, but also those where we don't see any, and learn a lot about genes along the way:
nature.com
Nature - A catalogue of predicted loss-of-function variants in 125,748 whole-exome and 15,708 whole-genome sequencing datasets from the Genome Aggregation Database (gnomAD) reveals the spectrum of...
5
149
431