Jungmann Lab
@JungmannLab
Followers
1K
Following
116
Media
47
Statuses
127
Our group at @LMU_Muenchen and @MPI_Biochem uses DNA nanotechnology to develop next-generation super-resolution microscopy techniques. #DNAPAINT
Munich, Germany
Joined May 2023
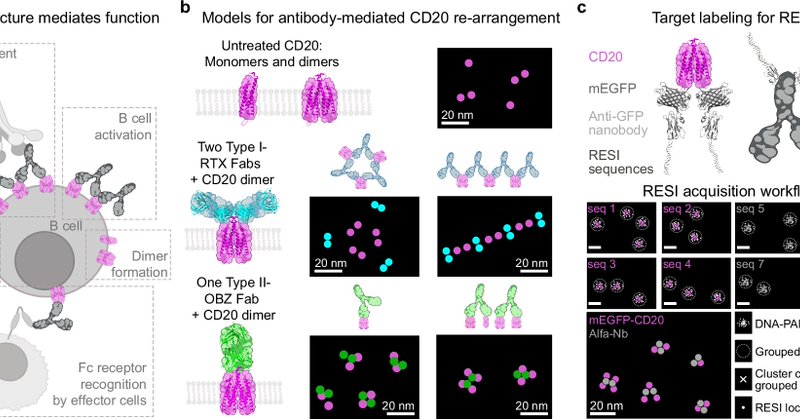
Congratulations to everyone involved: @IBaudrexel, @l_masu, @SusanneReinhar3, Jisoo Kwon, Ondřej Skořepa, Maite Llop, Sylvia Herter, Marina Bacac and Christian Klein. (6/6).Read the full story here:
nature.com
Nature Communications - The nanoscale organization of the antigen-antibody complexes influences the therapeutic action of monoclonal antibodies. Here, the authors present a multi-target 3D RESI...
1
2
3
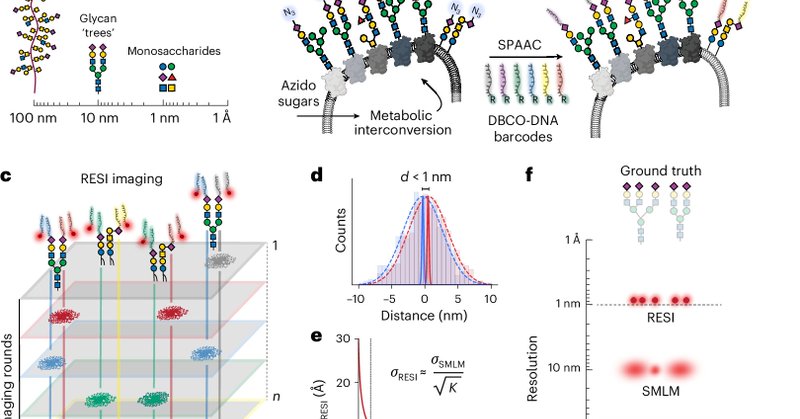
We're excited that the study is now out in Nature Nanotechnology @NatureNano
nature.com
Nature Nanotechnology - By combining bioorthogonal metabolic labelling and resolution enhancement through sequential imaging of DNA barcodes, the molecular organization of individual sugars in the...
ÅNGSTRÖM-RESOLUTION IMAGING OF CELL-SURFACE GLYCANS 🧬🎨🍬. The glycocalyx, our cells' sugar coat, holds secrets in immunology, cancer, viral infections, and more. Visualizing its molecular architecture was impossible… until now. #glycotime #microscopy.
1
21
99
RT @LMoeckl: Very happy to see this one out - the first molecular-resolution imaging of glycocalyx components in their native environment!….
0
5
0
RT @MPI_Biochem: Congratulations to Ralf on your election as a new EMBO member:. ❕Original press release from @EMBO: .
0
1
0
@MoniqueHonsa, @PhilippSteen, Larissa Heinze, Shuhan Xu, @heerpa, @IBaudrexel, @SusanneReinhar3, Ana Perovic, Jisoo Kwon, Ethan Oxley, Ross Dickins, @IkbenMaartje, @IanParish_AU.
0
0
2
Big congrats to @l_masu and @rafalkowalew who led the project as well as other co-authors that contributed to this work!!.
1
0
1
Spatial and stoichiometric in situ analysis of biomolecular oligomerization at single-protein resolution. We are excited to present our latest work published in @NatureComms .
nature.com
Nature Communications - Extracting quantitative information on biomolecular oligomerisation with high resolution remains a significant challenge. Here, the authors propose SPINNA, a framework that...
1
26
106
A huge thank you to all authors and collaborators whose dedication and teamwork made this protocol possible! @EduardUnterauer @EvaSchentarra @jekriste @MarrCarsten @FelipeOpaz_o @euforna.Let us know if you’re ready to generate your own single-cell spatial proteomics atlases! 🚀.
0
0
0