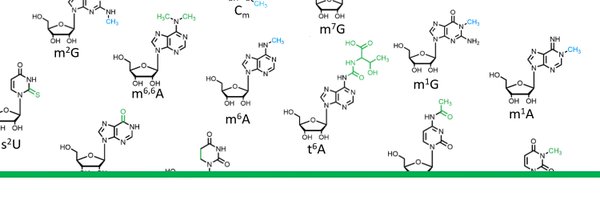
RMaP - TRR319
@ItsRMaP1
Followers
95
Following
3
Media
2
Statuses
21
All about "RNA Modifications and Processing - RMaP", the new TRR 319 funded by the DFG. Many exciting projects based in Mainz, Heidelberg, Mannheim and Dresden.
Joined June 2021
Just published - check out our exciting story -Queuosine‐tRNA promotes sex‐dependent learning and memory formation by maintaining codon‐biased translation elongation speed | The EMBO Journal https://t.co/3sjoUcfVlZ
9
32
111
with @clegrand17
@SchottJohanna
@DominickGuo
@AttheHelm3
@StoecklinUniHD
@schirmerlab
@dkfzEpigenetics
@ItsRMaP1
@embojournal
https://t.co/3sjoUcfVlZ
embopress.org
image image Queuosine (Q) is a modified nucleoside introduced in specific tRNAs by a complex comprising queuine tRNA ribosyltransferase 1 (QTRT1). Loss of Q‐tRNA modification by the knockout of Qtrt1...
0
3
3
We are very excited to announce that our new paper has been published in @Nature! Meet the new ‘RNAylation’ reaction: a viral ADP-ribosyltransferase (ART) is able to attach #RNA chains to E. coli proteins! Congratulations @JaeschkeLab @HoeferLab UrlaubLab https://t.co/A2ISBoXNLJ
3
47
185
Just one more month until our PhD students can enjoy lots of science, networking and refining their softskills at the RMaP student retreat in the baroque city of Rastatt, together with an interesting keynote lecture by @RuedaLab. Great job by our students organizing this event!👍
0
0
2
.@HFSP funding worth US$ 1.2 million for research project of @MonashUni, #UniMainz and @ETH_en investigating forms of #RNAmodification caused by #bacteria
https://t.co/BtEb2RdD06
#EffectorProteins #EukaryoticCells #epitranscriptome #Legionella #biomedicine @ItsRMaP1 @RMU_EMTHERA
0
1
3
Great news to announce for the TRR319 and the Helm group in particular: Research projects funded by the 'Human Frontier Science Program' to further investigate the many #RNA #modifications of #epitranscriptomics in an international collaboration.
0
0
5
🏆 Am 30.6. verleiht die GDCh den mit 7500 Euro dotierten Albrecht-Kossel-Preis an Andres Jäschke, Universität Heidelberg, für seine Arbeiten zur chemischen Biologie der Nukleinsäuren. Zur Pressemitteilung: https://t.co/szqQ5Sfq32
#gdchpreise Foto: Sabine Arndt
0
4
15
Commenting on a new type of RNA modification: Reversible internal phosphorylation of transfer RNA. Congratulations to the Suzuki lab for their great work! @ChemBiotech_UT
nature.com
Nature - Post-transcriptional modification of tRNA improves thermostability.
0
1
5
Thank you to all our speakers and guests from around the world who attended the RMaP & SPP1784 meeting last week. It was a great pleasure to finally see you all in person again for 4 days of #RNA science. We would also like to thank Tamaserv for sponsoring our evening sessions!
0
0
6
Happy to share our latest methodology article about "Normalized Ribo-Seq for Quantifying Absolute Global and Specific Changes in Translation". Congrats @SchottJohanna et al. https://t.co/yJQ62T0F8L
bio-protocol.org
New issues out! https://t.co/1sCLB5Lek7
#cancer
#neurosicences
#microbiology
#plantsci ...and more #protocols!
0
4
7
Happy to have contributed to this story with the first analysis of 2’O methylations in Xenopus by Ribomethseq ! Congrats to Jonathan Delhermite, @LafontaineLab and all co-authors.
Depletion of the ribosomal #RNA methyltransferase Fibrillarin leads to reduced eye size and severe craniofacial skeletal defects during early Xenopus embryogenesis #ribosome
https://t.co/Ss9qPNyY5X
@ULBruxelles
@ULBRecherche
@ULBSciences
@frsFNRS
@Dev_Gen_Lab
0
2
6
Join us at the 73rd Mosbacher Kolloquium: The world of #RNAs📢Deadline for early registration and abstract submission: January 31! GBM Young Members can apply for #TravelGrants at https://t.co/3bjorSdrW0 (see “information” and “grants”)
0
17
26
A surprise from a tiny #riboswitch: "Type I" riboswitch #ncRNAs for the modified nucleobase #preQ1 have been found by the @WedekindJoseph lab to stack two ligands in their #aptamer binding pocket. Only 34 nucleotides are used to achieve this RNA trick! https://t.co/uzTtCOVOMf
2
36
183
We offer a PhD position to investigate the role of #m6A in mammalian evolution as part of the #genevo network together with @KellerValsecchi and Susanne Foitzik, come and join us! https://t.co/CHXugBnE4n
0
15
18
Happy to share our work on “Balancing of mitochondrial translation through METTL8-mediated m3C modification of mitochondrial tRNAs” @MolecularCell. Congrats and thanks to Eva Schöller and all our collaborators for this excellent work.
2
13
63
Hi online world! Our lab finally made it to Twitter as well! We´re looking forward to sharing many lab stories and reading lots of fascinating research updates from around this planet. Sunset greetings from our office :) The Stoecklin Lab #research #Biochemistry
2
3
8
👇👇👇extended deadline October 31. I'm currently recruiting a PhD student as part of a newly funded consortium @ItsRMaP1 Check out details here: Interplay between tRNA splicing and tRNA modifications https://t.co/p1ZqBsyhd0
#hiring #tRNA #modifications with @JirkaPeschek Spread!
0
4
6
RMaP is hiring! The UniMed Heidelberg offers a part-time scientific project management position in the Integrated Research Training Group of RMaP (IRTG). Have a look below at the full desciption and apply until August 31st, 2021. #hiring #RNA 👇👇👇 https://t.co/T37C2A3DXW
0
5
1
An important matter! Please share the call @NatureGenet for direct #sequencing of full-length #RNAs to identify all #modifications! https://t.co/nrTRDzS8Zq
0
0
2