Ilya S
@IlyaSokolov314
Followers
13
Following
20
Media
0
Statuses
11
Postdoc at the Wellcome Sanger Institute, UK
Cambridge, England
Joined April 2023
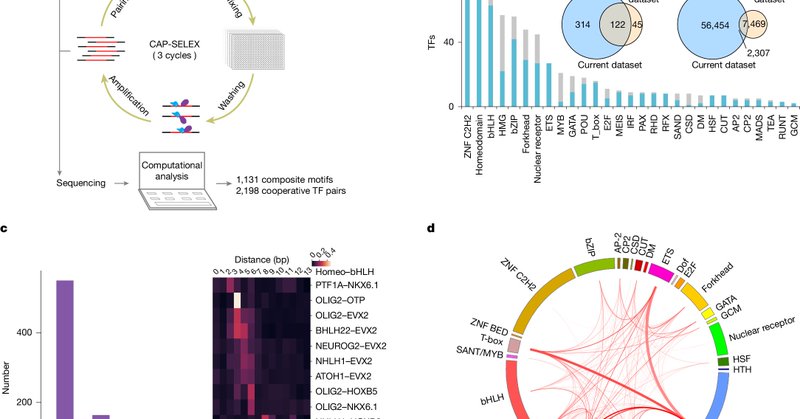
RT @Nature: Nature research paper: DNA-guided transcription factor interactions extend human gene regulatory code.
nature.com
Nature - A large-scale analysis of DNA-bound transcription factors (TFs) shows how the presence of DNA markedly affects the landscape of TF interactions, and identifies composite motifs that are...
0
15
0
@mariaosmala Special thanks to our supporting institutions: @Cambridge_Uni @Tongji_Uni @helsinkiuni @karolinskainst @UCIrvine,especially to all my amazing colleagues at the @sangerinstitute-and to Profs. Yimeng Yin & Jussi Taipale for conducting main experiments and inspiring leadership! 9/10.
1
0
1
Congratulations to Zhiyuan Xie, @mariaosmala, Ekaterina Morgunova and all co-authors on this achievement!.8/10.
1
0
1
Exciting news from our latest collaboration between Prof. Jussi Taipale's Lab Group ( and Prof. Yimeng Yin's Lab: our comprehensive screen of 58,000+ transcription factor pairs is now published in @Nature:.1/10.
nature.com
Nature - A large-scale analysis of DNA-bound transcription factors (TFs) shows how the presence of DNA markedly affects the landscape of TF interactions, and identifies composite motifs that are...
1
6
14