F. Kumara Mastrorosa
@fkmastrorosa
Followers
96
Following
279
Media
5
Statuses
89
Postdoctoral scholar in the @EichlerLab at @uwgenome. Interested in Mendelian disorders, long-read sequencing and structural variations. 🇮🇹 🇪🇺
Seattle, WA
Joined February 2019
Thank you, Dr. Danny Miller, for hosting, and fantastic job, Dr. @fkmastrorosa! https://t.co/yfTkbe081e
brotmanbaty.org
BBI
0
1
2
We updated our preprint describing chr21 centromere genetics/epigenetics in families with Down syndrome and general population! We found transgenerational methylation changes in a subset of families and that centromere size asymmetry is exclusive to T21! https://t.co/nLdsUzClyE
1
3
8
Catch Kati Buckingham from the Bamshad Lab at the #ASHG24 poster session this Thurs afternoon: “Variants in FGF20 underlie a novel breast malformation” Program ID: 2079T @bamshadmike @uwgenome
0
1
2
Check out CDR-Finder github for installation, testing, and a how-to-use guide:
github.com
Workflow to identify the centromere dip regions (CDR) from methyl BAMs - EichlerLab/CDR-Finder
0
0
1
Thanks to all the authors who contributed to the tool: @k0island, @AllisonRozanski, William Harvey, @EichlerLab, @glennis_logsdon
1
0
2
If you study centromeres and are interested in finding and visualizing the hypomethylated pockets where the kinetochore locates, check out CDR-Finder! The only inputs you need are a methyl-alignment, the centromere coordinates and your sample's assembly! https://t.co/ep94DIeYu9
1
6
9
Today I am presenting my study on centromere variations in Down syndrome families at #ASHG24! Come and see my poster in the Mendelian phenotypes section (Hall F) at 2:30pm!
1
1
3
Check out SVbyEye: A new visualization tool to characterize structural variation among whole-genome assemblies ( https://t.co/G0zGoHxZpe)
0
78
223
Great work from @LizziePlender! A deep dive into the secreted mucins MUC5A and MUC5B showing haplotype diversity (including VNTR variations!) and signatures of selection in the general population!
Check out our grad student Lizzie Plender’s 1st first-author paper “Structural and genetic diversity in the secreted mucins MUC5AC and MUC5B” using LRS to depict human genetic diversity, detect selection signatures, and develop genotyping strategies. https://t.co/5IpavkgSjq
0
0
3
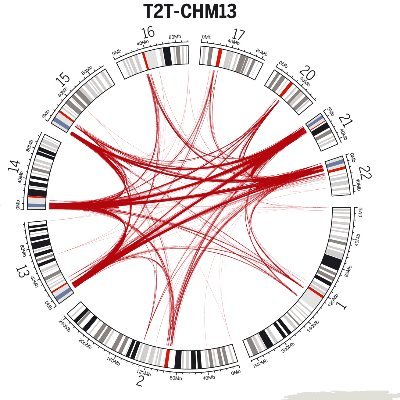
I'm absolutely thrilled to share that our work on human centromere variation and evolution has been published in @Nature! We completely sequenced all centromeres from a second human genome & compared them to those from the first complete human genome, uncovering new sequences,
14
136
525
Long live the long-read sequencing (LRS) technology! The genomics field's new superpower to read even the darkest, repetitive parts--centromeres and telomeres--of the human genome is beginning to solve decades to centuries-old genetic puzzles, one by one. Last year, we saw how
A potential role for centromere dysfunction in Down syndrome! We generated the first complete sequence of all three chr21 centromeres in an individual with Down syndrome & found that one centromere is ~11-fold smaller than usual and another has an atypical epigenetic landscape!
0
11
56
Thanks to all the people that worked on the project: @AllisonRozanski, William Harvey, Jordan Knuth, Gage Garcia, @zhaneel779 @kendra_hoekzema
0
0
1
Happy to share our work on Down syndrome! We sequenced and assembled all chr21 centromeres of a parent-proband trio with Trisomy 21 (MMIE) revealing centromere asymmetry and differences in kinetochore attachment @glennis_logsdon @EichlerLab @EichlerEE
https://t.co/APw43tQH9n
1
8
45
It is unfortunate that this has to come out on the 7th day of the UW Postdocs (myself included) and RSE strike, battling for fair contracts and living wages! @UAW4121
Our review on current and future applications of long-read sequencing for the study of Mendelian conditions has just been published! Technologies, inputs, costs, and workflows are discussed in this paper! @danrdanny @EichlerLab
https://t.co/t84XiLoxsL
0
3
6
Our review on current and future applications of long-read sequencing for the study of Mendelian conditions has just been published! Technologies, inputs, costs, and workflows are discussed in this paper! @danrdanny @EichlerLab
https://t.co/t84XiLoxsL
genomemedicine.biomedcentral.com
Advances in clinical genetic testing, including the introduction of exome sequencing, have uncovered the molecular etiology for many rare and previously unsolved genetic disorders, yet more than half...
0
15
10
Excited to announce the first view of human centromere variation! We completely sequenced all centromeres from a second human genome & compared them to those from the first complete human genome, uncovering new sequences, structures, & chromatin landscapes https://t.co/FdWmSYCV6j
16
217
826