Elena Vasileva
@EVasilevaSci
Followers
339
Following
825
Media
57
Statuses
243
K99/R00 NIH NCI Postdoctoral Fellow at @ChildrensLA. I use #zebrafish 🐟 to learn more about human cancer #EwingSarcoma @AmatrudaLab
Los Angeles, CA
Joined February 2019
Applications are still open! Expand your data analysis skills during our Advanced #SingleCell RNA-Seq Workshop for #PediatricCancer researchers. 📆 December 8-12, 2025 💻 Virtual ⬇️ Learn more and apply! https://t.co/7jJAoFUczU
ccdatalab.org
Applications are open for the Data Lab's upcoming workshop, which will cover advanced topics in the analysis of single-cell RNA-seq data for researchers studying pediatric cancer. The course will be...
0
1
2
Hey everyone! @zdmsECI is excited to showcase our #zdm speakers. First up is Elena Vasileva. She will be giving a talk titled “Development of Ewing Sarcoma Through Embryonic Reprogramming of Neural Crest to Mesoderm”. @ZDMSociety
0
4
5
9/9 We hope this model will deepen our understanding of the origins of human #EwingSarcoma and will speed the development of better therapies. @amatrudalab.bsky.social
0
0
0
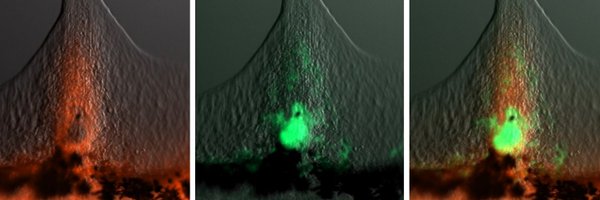
8/9. Strikingly, EWSR1::FLI1+ cells induced ectopic fin formation by locally activating Fgfr and Hox signaling, eventually giving rise to tumors. This suggests that pre-tumor cells hijack developmental programs to establish a local signaling environment for tumor initiation.
1
0
0
7/9 By integrating chromatin accessibility with genome-wide EWSR1::FLI1 binding profiles, we show that the fusion reprograms cells by binding ETS motif-containing developmental enhancers. We also observed EWSR1::FLI1 binding to GGAA repeats, consistent with human data.
1
0
0
6/9 Single-cell transcriptomic and chromatin profiling of zebrafish tumors revealed that EWSR1::FLI1 expression in neural crest cells drives a mesenchymal/mesodermal shift, with mesodermal genes like #Brachyury (T) strongly upregulated. #NCCs #mesoderm #reprogramming
1
0
0
5/9. Next, we generated a stable zebrafish line with tissue-specific expression of human EWSR1::FLI1 in NCCs. This led to the development of pre-tumor outgrowths and, eventually, massive tumors, demonstrating that the expression of the oncofusion in NCCs is tumorigenic in vivo.
1
0
0
4/9 Using our mosaic zebrafish model of Ewing sarcoma, we traced EWSR1-FLI1 positive cells in early development and found that lineage, timing, and expression level of the oncofusion are critical. In our model, neural crest cells uniquely tolerated this toxic oncofusion in vivo.
1
0
0
3/9 In this study, we use zebrafish genetic cancer models to investigate the origin of #EwingSarcoma. We show that EWSR1::FLI1 alters embryonic cell fate decisions, reprogramming neural crest cells into a mesoderm-like state that initiates this devastating childhood cancer.
1
0
0
2/9 Huge thanks to Dr. Arata @clairearataphd and Dr. Crump @CrumpLab, and the amazing @ChildrensLA environment that made this work possible. Very grateful to my mentor, Dr. Amatruda, and our fantastic Lab.
1
0
0
1/9 Thrilled to share that our paper, “Origin of Ewing sarcoma by embryonic reprogramming of neural crest to mesoderm,” is now published in @CellReports! @amatrudalab.bsky.social @CrumpLab
https://t.co/Eioj5M6Nrr
cell.com
Using a zebrafish genetic model of Ewing sarcoma, Vasileva et al. provide evidence for a neural crest origin of the disease. These findings offer new insight into how a single oncogenic fusion can...
1
1
0
Many thanks to @CancerDataLab for the valuable support, training and resources.
In our latest blog post, we’re highlighting a few researchers who have drawn on the Data Lab’s resources and expertise in their remarkable efforts to accelerate new treatment and cure discovery! #ChildhoodCancerAwarenessMonth
https://t.co/MuMZGbk5zc
0
0
1
Check out this impressive new study from the Grünewald Lab, revealing a novel way to target Ewing sarcoma cells via CXADR-mediated AKT signaling. Congratulations to Alina Ritter and Thomas Grünewald! https://t.co/pwRDDbjxNe
0
0
1
We’re hiring! 🚨 My lab at @UUtah @UofUMandI is looking for a Senior Laboratory Specialist to join our research on glycobiology, metabolism, and inflammation. Apply here 👇 https://t.co/2ZH684Z8xk
#Glycotime #Hiring #ResearchCareers
0
4
10
Here is our new review article: “The role of microheterogeneity in cell fate decisions in neural progenitors and neural crest” https://t.co/7EYuPJaCE2 Check it out if you are curious about how cell cycle or transcriptional noise affect fate selection in cell lineages.
0
7
20
Zebrafish people! The @ZDMSociety annual meeting is in Boston this year, with a great lineup. Abstract submission is closing May 27th. Go register now for this always fun conference #zebrafish
https://t.co/2QBggT55G5
0
2
12
Congratulations to @JackKucinski and @Chriskuo_MD on their @AACR Scholar-in-Training Awards! @KendallLabNCH @amatrudaLab
0
1
8
I highly recommend these excellent workshops on data analysis. A big thank you to @CancerDataLab for doing such a great job – it is incredibly valuable to our pediatric community.
Hear from Drs. Eric Wafula and Elena Vasileva who sat down with us to share their experiences at Data Lab training workshops! Learn why they believe our #SingleCell #RNASeq courses are essential for #PediatricCancer researchers. https://t.co/zOShGEMB7D
0
0
2
Just completed the @AlexsLemonade Childhood Cancer Data Lab Advanced Single-Cell RNA-Seq Workshop. This was the second time I’ve attended the @CancerDataLab workshop. So grateful for this wonderful opportunity. Huge thanks to our amazing instructors -Stephanie Spielman, Jen
0
0
11