DePace Lab
@DePaceLab
Followers
1K
Following
516
Media
4
Statuses
2K
A reminder that my department at HMS is hiring! Applications due Oct. 31st. Its a great place to work, and we're excited about finding our next new colleague.
.@HMS_SysBio is seeking an Assistant Professor using quantitative experimental, computational, synthetic and/or theoretical approaches. Please apply at 👇🏿 https://t.co/OKBag8cPol 📆 Oct 31st #SystemsBiology #SysBioJobs #ScienceJobs
0
7
4
Thrilled to share our new (unredacted) report in @sciencemagazine with evidence for cooperation influencing outcomes! Led by Caleb Bashor & @nikitpatels, we built cooperative TF assemblies and showed how they enable complex signal processing in cells:
4
38
138
Excited to share new work from @ClarissaScholes! She measured how 2 Kr shadow enhancers combine their output using live imaging. Take home: 1 + 1 sometimes is a lot less than 2, sometimes only a little bit less. Output is not a simple function of enhancer expression level.
0
13
23
Excited that our review on the MZT in Drosophila is now online at Open Biology ( https://t.co/Xv5x7rciCo). Great work from recent PhD grad Danielle Hamm.
0
8
32
Undergraduates -- apply for a Summer Internship in Systems Biology! You get to explore Harvard and do cool research while being paid.
2
13
6
Don't forget to apply to join the fabulous group of interdisciplinary students and faculty in the Harvard PhD program in Systems Biology! Deadline Dec 1. No GRE needed, application fees can be waived, stipend, tuition and health insurance included.
0
6
7
@FORsymp Happy to say that we will be matching the next $1000 of donations to @FORsymp! Everyone wants the standard of mentoring in academia to improve, this is a great way to start driving change. #GivingTuesday
1
4
10
more evidence for the functional role of Zelda hubs in mediating transcription factor binding to their targets. Really exciting to see this line of research blossoming!
0
7
20
Hello #ABRCMS2018 ! Come talk to us about the Harvard PhD Program in Systems Biology! We’re in the Exhibit Hall booth 812.
0
5
8
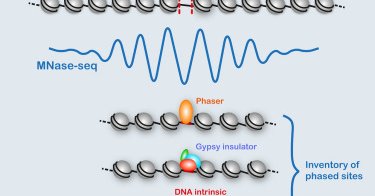
1/ Our work on phased nucleosome arrays in the Drosophila genome is out: https://t.co/HLNbdDSI1k In it, we used spectral analysis to map phased arrays throughout the genome.
cell.com
Patches of evenly spaced nucleosomes mark regulatory elements in the genome. Baldi et al. comprehensively map such phased arrays throughout the Drosophila genome. By reconstituting phased arrays in a...
3
13
20
The role of chromatin accessibility in cis-regulatory evolution
0
2
8
Happy to finally share this (long in progress) work with the world. Lots of hard work by @gizemkly (especially), @JLachowiec, and Ulises Rosas. Redundant and cryptic enhancer activities of the Drosophila yellow gene
2
25
33
It's on bioarxiv finally! In this paper we dissect a GRN into dynamical modules and use them to study the networks evovability. Give us your feedback too! It hasn't been submitted yet. @yoginho
3
26
67
Interesting study from @DePaceLab
https://t.co/9L2gdZ7E6s Revisiting a 1989 classic from Dreiver et al https://t.co/vSOqL6ZBGc by applying quant analysis to test whether gene expression pattern controlled by Bicoid binding affinity turns out it's a little more complicated
nature.com
Nature - Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding sites for the bicoid morphogen
0
4
8
"Our results emphasize that the bacterial view of transcription regulation, where pairwise interactions between regulatory proteins dominate, must be reexamined in animals..."
Excited to share new work from @JPbiophysics & the Gunawardena lab. Mutants, knockdowns & modeling to understand how a classic Drosophila enhancer drives sharp gene expression. Spoiler : its not simple cooperative DNA binding by TFs. https://t.co/AR3MKg5RxN
0
3
3
Excited to share new work from @JPbiophysics & the Gunawardena lab. Mutants, knockdowns & modeling to understand how a classic Drosophila enhancer drives sharp gene expression. Spoiler : its not simple cooperative DNA binding by TFs. https://t.co/AR3MKg5RxN
0
20
44
Excited to share this work from @ClarissaScholes - lots left to learn about how shadow enhancers collaborate to control gene expression. She took an important first step - coming to grips with the computation.
0
8
19